A unified model of protein dynamics
- PMID: 19251640
- PMCID: PMC2649210
- DOI: 10.1073/pnas.0900336106
A unified model of protein dynamics
Abstract
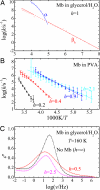
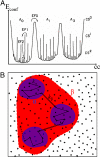
Protein functions require conformational motions. We show here that the dominant conformational motions are slaved by the hydration shell and the bulk solvent. The protein contributes the structure necessary for function. We formulate a model that is based on experiments, insights from the physics of glass-forming liquids, and the concepts of a hierarchically organized energy landscape. To explore the effect of external fluctuations on protein dynamics, we measure the fluctuations in the bulk solvent and the hydration shell with broadband dielectric spectroscopy and compare them with internal fluctuations measured with the Mössbauer effect and neutron scattering. The result is clear. Large-scale protein motions are slaved to the fluctuations in the bulk solvent. They are controlled by the solvent viscosity, and are absent in a solid environment. Internal protein motions are slaved to the beta fluctuations of the hydration shell, are controlled by hydration, and are absent in a dehydrated protein. The model quantitatively predicts the rapid increase of the mean-square displacement above approximately 200 K, shows that the external beta fluctuations determine the temperature- and time-dependence of the passage of carbon monoxide through myoglobin, and explains the nonexponential time dependence of the protein relaxation after photodissociation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

