Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin
- PMID: 19251641
- PMCID: PMC2657435
- DOI: 10.1073/pnas.0813360106
Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin
Abstract
Protein secretion is a common property of pathogenic microbes. Gram-negative bacterial pathogens use at least 6 distinct extracellular protein secretion systems to export proteins through their multilayered cell envelope and in some cases into host cells. Among the most widespread is the newly recognized Type VI secretion system (T6SS) which is composed of 15-20 proteins whose biochemical functions are not well understood. Using crystallographic, biochemical, and bioinformatic analyses, we identified 3 T6SS components, which are homologous to bacteriophage tail proteins. These include the tail tube protein; the membrane-penetrating needle, situated at the distal end of the tube; and another protein associated with the needle and tube. We propose that T6SS is a multicomponent structure whose extracellular part resembles both structurally and functionally a bacteriophage tail, an efficient machine that translocates proteins and DNA across lipid membranes into cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
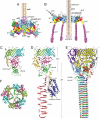
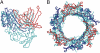
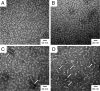

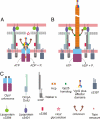
Comment in
-
Structural similarity of tailed phages and pathogenic bacterial secretion systems.Proc Natl Acad Sci U S A. 2009 Mar 17;106(11):4067-8. doi: 10.1073/pnas.0901205106. Epub 2009 Mar 10. Proc Natl Acad Sci U S A. 2009. PMID: 19276114 Free PMC article. Review. No abstract available.
References
-
- Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: A beginner's guide. Curr Opin Microbiol. 2008;11:3–8. - PubMed
-
- Folkesson A, Lofdahl S, Normark S. The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol. 2002;153:537–545. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

