Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing
- PMID: 19251664
- PMCID: PMC2657401
- DOI: 10.1073/pnas.0810666106
Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing
Abstract
In search for physiological pathways affecting alternative splicing through its kinetic coupling with transcription, we found that membrane depolarization of neuronal cells triggers the skipping of exon 18 from the neural cell adhesion molecule (NCAM) mRNA, independently of the calcium/calmodulin protein kinase IV pathway. We show that this exon responds to RNA polymerase II elongation, because its inclusion is increased by a slow polymerase II mutant. Depolarization affects the chromatin template in a specific way, by causing H3K9 hyper-acetylation restricted to an internal region of the NCAM gene surrounding the alternative exon. This intragenic histone hyper-acetylation is not paralleled by acetylation at the promoter, is associated with chromatin relaxation, and is linked to H3K36 tri-methylation. The effects on acetylation and splicing fully revert when the depolarizing conditions are withdrawn and can be both duplicated and potentiated by the histone deacetylase inhibitor trichostatin A. Our results are consistent with a mechanism involving the kinetic coupling of splicing and transcription in response to depolarization through intragenic epigenetic changes on a gene that is relevant for the differentiation and function of neuronal cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


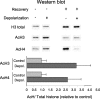

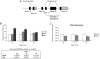
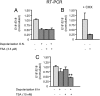
Similar articles
-
Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation.EMBO J. 2013 Aug 14;32(16):2264-74. doi: 10.1038/emboj.2013.167. Epub 2013 Jul 26. EMBO J. 2013. PMID: 23892457 Free PMC article.
-
Chromatin and alternative splicing.Cold Spring Harb Symp Quant Biol. 2010;75:103-11. doi: 10.1101/sqb.2010.75.023. Epub 2011 Feb 2. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21289049
-
Histone hyperacetylation and exon skipping: a calcium-mediated dynamic regulation in cardiomyocytes.Nucleus. 2015;6(4):273-8. doi: 10.1080/19491034.2015.1081324. Nucleus. 2015. PMID: 26325491 Free PMC article.
-
A saga of cancer epigenetics: linking epigenetics to alternative splicing.Biochem J. 2017 Mar 7;474(6):885-896. doi: 10.1042/BCJ20161047. Biochem J. 2017. PMID: 28270561 Review.
-
Coupling transcription and alternative splicing.Adv Exp Med Biol. 2007;623:175-89. doi: 10.1007/978-0-387-77374-2_11. Adv Exp Med Biol. 2007. PMID: 18380347 Review.
Cited by
-
A Broad Set of Chromatin Factors Influences Splicing.PLoS Genet. 2016 Sep 23;12(9):e1006318. doi: 10.1371/journal.pgen.1006318. eCollection 2016 Sep. PLoS Genet. 2016. PMID: 27662573 Free PMC article.
-
Getting up to speed with transcription elongation by RNA polymerase II.Nat Rev Mol Cell Biol. 2015 Mar;16(3):167-77. doi: 10.1038/nrm3953. Epub 2015 Feb 18. Nat Rev Mol Cell Biol. 2015. PMID: 25693130 Free PMC article. Review.
-
Alternative splicing: a pivotal step between eukaryotic transcription and translation.Nat Rev Mol Cell Biol. 2013 Mar;14(3):153-65. doi: 10.1038/nrm3525. Epub 2013 Feb 6. Nat Rev Mol Cell Biol. 2013. PMID: 23385723 Review.
-
Histone deacetylase inhibitors improve antisense-mediated exon-skipping efficacy in mdx mice.Mol Ther Nucleic Acids. 2022 Nov 21;30:606-620. doi: 10.1016/j.omtn.2022.11.017. eCollection 2022 Dec 13. Mol Ther Nucleic Acids. 2022. PMID: 36514350 Free PMC article.
-
Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition.Cell Res. 2013 Nov;23(11):1256-69. doi: 10.1038/cr.2013.110. Epub 2013 Aug 13. Cell Res. 2013. PMID: 23938295 Free PMC article.
References
-
- Proudfoot NJ, et al. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. - PubMed
-
- Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–3871. - PubMed
-
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. - PubMed
-
- de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

