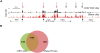
Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns
- PMID: 19251738
- PMCID: PMC2665778
- DOI: 10.1101/gr.086983.108
Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns
Abstract
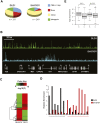
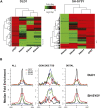
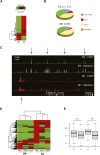
CHD7 is a member of the chromodomain helicase DNA binding domain family of ATP-dependent chromatin remodeling enzymes. De novo mutation of the CHD7 gene is a major cause of CHARGE syndrome, a genetic disease characterized by a complex constellation of birth defects (Coloboma of the eye, Heart defects, Atresia of the choanae, severe Retardation of growth and development, Genital abnormalities, and Ear abnormalities). To gain insight into the function of CHD7, we mapped the distribution of the CHD7 protein on chromatin using the approach of chromatin immunoprecipitation on tiled microarrays (ChIP-chip). These studies were performed in human colorectal carcinoma cells, human neuroblastoma cells, and mouse embryonic stem (ES) cells before and after differentiation into neural precursor cells. The results indicate that CHD7 localizes to discrete locations along chromatin that are specific to each cell type, and that the cell-specific binding of CHD7 correlates with a subset of histone H3 methylated at lysine 4 (H3K4me). The CHD7 sites change concomitantly with H3K4me patterns during ES cell differentiation, suggesting that H3K4me is part of the epigenetic signature that defines lineage-specific association of CHD7 with specific sites on chromatin. Furthermore, the CHD7 sites are predominantly located distal to transcription start sites, most often contained within DNase hypersensitive sites, frequently conserved, and near genes expressed at relatively high levels. These features are similar to those of gene enhancer elements, raising the possibility that CHD7 functions in enhancer mediated transcription, and that the congenital anomalies in CHARGE syndrome are due to alterations in transcription of tissue-specific genes normally regulated by CHD7 during development.
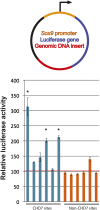
Figures


References
-
- Aramaki M., Udaka T., Kosaki R., Makita Y., OkamAto N., Yoshihashi H., Oki H., Nanao K., Moriyama N., Oku S., et al. Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J. Pediatr. 2006;148:410–414. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Blake K.D., Davenport S.L., Hall B.D., Hefner M.A., Pagon R.A., Williams M.S., Lin A.E., Graham J.M., Jr CHARGE association: An update and review for the primary pediatrician. Clin. Pediatr. (Phila.) 1998;37:159–173. - PubMed
-
- Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006;16:656–668. - PMC - PubMed
-
- Bosman E.A., Penn A.C., Ambrose J.C., Kettleborough R., Stemple D.L., Steel K.P. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
