Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes
- PMID: 19251817
- PMCID: PMC2683692
- DOI: 10.1124/dmd.109.026765
Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes
Abstract
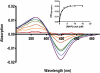
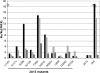
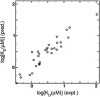
Although the human lung cytochrome P450 2A13 (CYP2A13) and its liver counterpart cytochrome P450 2A6 (CYP2A6) are 94% identical in amino acid sequence, they metabolize a number of substrates with substantially different efficiencies. To determine differences in binding for a diverse set of cytochrome P450 2A ligands, we have measured the spectral binding affinities (K(D)) for nicotine, phenethyl isothiocyanate (PEITC), coumarin, 2'-methoxyacetophenone (MAP), and 8-methoxypsoralen. The differences in the K(D) values for CYP2A6 versus CYP2A13 ranged from 74-fold for 2'-methoxyacetophenone to 1.1-fold for coumarin, with CYP2A13 demonstrating the higher affinity. To identify active site amino acids responsible for the differences in binding of MAP, PEITC, and coumarin, 10 CYP2A13 mutant proteins were generated in which individual amino acids from the CYP2A6 active site were substituted into CYP2A13 at the corresponding position. Titrations revealed that substitutions at positions 208, 300, and 301 individually had the largest effects on ligand binding. The collective relevance of these amino acids to differential ligand selectivity was verified by evaluating binding to CYP2A6 mutant enzymes that incorporate several of the CYP2A13 amino acids at these positions. Inclusion of four CYP2A13 amino acids resulted in a CYP2A6 mutant protein (I208S/I300F/G301A/S369G) with binding affinities for MAP and PEITC much more similar to those observed for CYP2A13 than to those for CYP2A6 without altering coumarin binding. The structure-based quantitative structure-activity relationship analysis using COMBINE successfully modeled the observed mutant-ligand trends and emphasized steric roles for active site residues including four substituted amino acids and an adjacent conserved Leu(370).
Figures


References
-
- Bao Z, He XY, Ding X, Prabhu S, and Hong JY (2005) Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab Dispos 33 258–261. - PubMed
-
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, and Karplus M (1983) CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J Comp Chem 4 187–217.
-
- DeLano WL (2003) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA.
-
- Fernandez-Salguero P, Hoffman SM, Cholerton S, Mohrenweiser H, Raunio H, Rautio A, Pelkonen O, Huang JD, Evans WE, and Idle JR (1995) A genetic-polymorphism in coumarin 7-hydroxylation—sequence of the human Cyp2a genes and identification of variant Cyp2a6 alleles. Am J Hum Genet 57 651–660. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

