Phylogeography of Francisella tularensis: global expansion of a highly fit clone
- PMID: 19251856
- PMCID: PMC2668398
- DOI: 10.1128/JB.01786-08
Phylogeography of Francisella tularensis: global expansion of a highly fit clone
Abstract
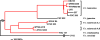
Francisella tularensis contains several highly pathogenic subspecies, including Francisella tularensis subsp. holarctica, whose distribution is circumpolar in the northern hemisphere. The phylogeography of these subspecies and their subclades was examined using whole-genome single nucleotide polymorphism (SNP) analysis, high-density microarray SNP genotyping, and real-time-PCR-based canonical SNP (canSNP) assays. Almost 30,000 SNPs were identified among 13 whole genomes for phylogenetic analysis. We selected 1,655 SNPs to genotype 95 isolates on a high-density microarray platform. Finally, 23 clade- and subclade-specific canSNPs were identified and used to genotype 496 isolates to establish global geographic genetic patterns. We confirm previous findings concerning the four subspecies and two Francisella tularensis subsp. tularensis subpopulations and identify additional structure within these groups. We identify 11 subclades within F. tularensis subsp. holarctica, including a new, genetically distinct subclade that appears intermediate between Japanese F. tularensis subsp. holarctica isolates and the common F. tularensis subsp. holarctica isolates associated with the radiation event (the B radiation) wherein this subspecies spread throughout the northern hemisphere. Phylogenetic analyses suggest a North American origin for this B-radiation clade and multiple dispersal events between North America and Eurasia. These findings indicate a complex transmission history for F. tularensis subsp. holarctica.
Figures

References
-
- Achtman, M., G. Morelli, P. Zhu, T. Wirth, I. Diehl, B. Kusecek, A. J. Vogler, D. M. Wagner, C. J. Allender, W. R. Easterday, V. Chenal-Francisque, P. Worsham, N. R. Thomson, J. Parkhill, L. E. Lindler, E. Carniel, and P. Keim. 2004. Microevolution and history of the plague bacillus, Yersinia pestis. Proc. Natl. Acad. Sci. USA 10117837-17842. - PMC - PubMed
-
- Auerbach, R. K. 2006. sDACS: a novel in silico SNP discovery and classification method for bacterial pathogens. Northern Arizona University, Flagstaff, AZ.
-
- Beckstrom-Sternberg, S. M., R. K. Auerbach, S. Godbole, J. V. Pearson, J. S. Beckstrom-Sternberg, Z. Deng, C. Munk, K. Kubota, Y. Zhou, D. Bruce, J. Noronha, R. H. Scheuermann, A. Wang, X. Wei, J. Wang, J. Hao, D. M. Wagner, T. S. Brettin, N. Brown, P. Gilna, and P. S. Keim. 2007. Complete genomic characterization of a pathogenic A.II strain of Francisella tularensis subspecies tularensis. PLoS ONE 2e947. - PMC - PubMed
-
- Centers for Disease Control and Prevention, Office of Inspector General, Department of Health and Human Services (HHS). 2005. Possession, use, and transfer of select agents and toxins. Final rule. Fed. Regist. 7013293-13325. - PubMed
-
- Cha, R. S., H. Zarbl, P. Keohavong, and W. G. Thilly. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl. 214-20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

