Identification and analyses of candidate genes for rpp4-mediated resistance to Asian soybean rust in soybean
- PMID: 19251904
- PMCID: PMC2675740
- DOI: 10.1104/pp.108.134551
Identification and analyses of candidate genes for rpp4-mediated resistance to Asian soybean rust in soybean
Abstract
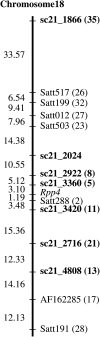
Asian soybean rust is a formidable threat to soybean (Glycine max) production in many areas of the world, including the United States. Only five sources of resistance have been identified (Resistance to Phakopsora pachyrhizi1 [Rpp1], Rpp2, Rpp3, Rpp4, and Rpp5). Rpp4 was previously identified in the resistant genotype PI459025B and mapped within 2 centimorgans of Satt288 on soybean chromosome 18 (linkage group G). Using simple sequence repeat markers, we developed a bacterial artificial chromosome contig for the Rpp4 locus in the susceptible cv Williams82 (Wm82). Sequencing within this region identified three Rpp4 candidate disease resistance genes (Rpp4C1-Rpp4C3 [Wm82]) with greatest similarity to the lettuce (Lactuca sativa) RGC2 family of coiled coil-nucleotide binding site-leucine rich repeat disease resistance genes. Constructs containing regions of the Wm82 Rpp4 candidate genes were used for virus-induced gene silencing experiments to silence resistance in PI459025B, confirming that orthologous genes confer resistance. Using primers developed from conserved sequences in the Wm82 Rpp4 candidate genes, we identified five Rpp4 candidate genes (Rpp4C1-Rpp4C5 [PI459025B]) from the resistant genotype. Additional markers developed from the Wm82 Rpp4 bacterial artificial chromosome contig further defined the region containing Rpp4 and eliminated Rpp4C1 (PI459025B) and Rpp4C3 (PI459025B) as candidate genes. Sequencing of reverse transcription-polymerase chain reaction products revealed that Rpp4C4 (PI459025B) was highly expressed in the resistant genotype, while expression of the other candidate genes was nearly undetectable. These data support Rpp4C4 (PI459025B) as the single candidate gene for Rpp4-mediated resistance to Asian soybean rust.
Figures


Similar articles
-
The Soybean Rpp3 Gene Encodes a TIR-NBS-LRR Protein that Confers Resistance to Phakopsora pachyrhizi.Mol Plant Microbe Interact. 2024 Jul;37(7):561-570. doi: 10.1094/MPMI-01-24-0007-R. Epub 2024 Jul 13. Mol Plant Microbe Interact. 2024. PMID: 38569009
-
Fine mapping of the Asian soybean rust resistance gene Rpp2 from soybean PI 230970.Theor Appl Genet. 2015 Mar;128(3):387-96. doi: 10.1007/s00122-014-2438-0. Epub 2014 Dec 12. Theor Appl Genet. 2015. PMID: 25504467
-
Transcriptome analyses and virus induced gene silencing identify genes in the Rpp4-mediated Asian soybean rust resistance pathway.Funct Plant Biol. 2013 Oct;40(10):1029-1047. doi: 10.1071/FP12296. Funct Plant Biol. 2013. PMID: 32481171
-
SNP assay to detect the 'Hyuuga' red-brown lesion resistance gene for Asian soybean rust.Theor Appl Genet. 2010 Oct;121(6):1023-32. doi: 10.1007/s00122-010-1368-8. Epub 2010 Jun 8. Theor Appl Genet. 2010. PMID: 20532750 Free PMC article.
-
Molecular mapping of two loci that confer resistance to Asian rust in soybean.Theor Appl Genet. 2008 Jun;117(1):57-63. doi: 10.1007/s00122-008-0752-0. Epub 2008 Apr 8. Theor Appl Genet. 2008. PMID: 18392802
Cited by
-
Evolutionary and comparative analyses of the soybean genome.Breed Sci. 2012 Jan;61(5):437-44. doi: 10.1270/jsbbs.61.437. Epub 2012 Feb 4. Breed Sci. 2012. PMID: 23136483 Free PMC article.
-
Molecular Breeding to Overcome Biotic Stresses in Soybean: Update.Plants (Basel). 2022 Jul 28;11(15):1967. doi: 10.3390/plants11151967. Plants (Basel). 2022. PMID: 35956444 Free PMC article. Review.
-
RNA-Seq and Comparative Transcriptomic Analyses of Asian Soybean Rust Resistant and Susceptible Soybean Genotypes Provide Insights into Identifying Disease Resistance Genes.Int J Mol Sci. 2023 Aug 30;24(17):13450. doi: 10.3390/ijms241713450. Int J Mol Sci. 2023. PMID: 37686258 Free PMC article.
-
An pair of an atypical NLR encoding genes confer Asian soybean rust resistance in soybean.Nat Commun. 2024 Apr 17;15(1):3310. doi: 10.1038/s41467-024-47611-y. Nat Commun. 2024. PMID: 38632249 Free PMC article.
-
A genome-wide association study and genomic prediction for Phakopsora pachyrhizi resistance in soybean.Front Plant Sci. 2023 May 29;14:1179357. doi: 10.3389/fpls.2023.1179357. eCollection 2023. Front Plant Sci. 2023. PMID: 37313252 Free PMC article.
References
-
- Abbott JC, Aanensen DM, Rutherford K, Butcher S, Spratt BG (2005) WebACT: an online companion for the Artemis Comparison Tool. Bioinformatics 21 3665–3666 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

