A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana
- PMID: 19252080
- PMCID: PMC2660620
- DOI: 10.1105/tpc.108.064535
A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana
Abstract
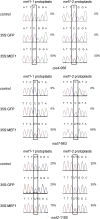
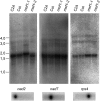
RNA editing in flowering plant mitochondria alters 400 to 500 nucleotides from C to U, changing the information content of most mRNAs and some tRNAs. So far, none of the specific or general factors responsible for RNA editing in plant mitochondria have been identified. Here, we characterize a nuclear-encoded gene that is involved in RNA editing of three specific sites in different mitochondrial mRNAs in Arabidopsis thaliana, editing sites rps4-956, nad7-963, and nad2-1160. The encoded protein MITOCHONDRIAL RNA EDITING FACTOR1 (MEF1) belongs to the DYW subfamily of pentatricopeptide repeat proteins. Amino acid identities altered in MEF1 from ecotype C24, in comparison to Columbia, lower the activity at these editing sites; single amino acid changes in mutant plants inactivate RNA editing. These variations most likely modify the affinity of the editing factor to the affected editing sites in C24 and in the mutant plants. Since lowered and even absent RNA editing is tolerated at these sites, the amino acid changes may be silent for the respective protein functions. Possibly more than these three identified editing sites are addressed by this first factor identified for RNA editing in plant mitochondria.
Figures




References
-
- Andrés, C., Lurin, C., and Small, I.D. (2007). The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant. 129 14–22.
-
- Chateigner-Boutin, A.L., Ramos-Vega, M., Guevara-García, A., Andrés, C., de la Luz Gutiérrez-Nava, M., Cantero, A., Delannoy, E., Jiménez, L.F., Lurin, C., Small, I., and León, P. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56 590–602. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

