The RPN5 subunit of the 26s proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis
- PMID: 19252082
- PMCID: PMC2660617
- DOI: 10.1105/tpc.108.064444
The RPN5 subunit of the 26s proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis
Abstract
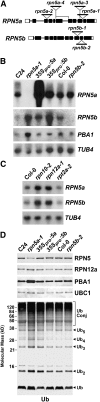
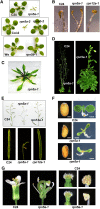
The 26S proteasome is an essential multicatalytic protease complex that degrades a wide range of intracellular proteins, especially those modified with ubiquitin. Arabidopsis thaliana and other plants use pairs of genes to encode most of the core subunits, with both of the isoforms often incorporated into the mature complex. Here, we show that the gene pair encoding the regulatory particle non-ATPase subunit (RPN5) has a unique role in proteasome function and Arabidopsis development. Homozygous rpn5a rpn5b mutants could not be generated due to a defect in male gametogenesis. While single rpn5b mutants appear wild-type, single rpn5a mutants display a host of morphogenic defects, including abnormal embryogenesis, partially deetiolated development in the dark, a severely dwarfed phenotype when grown in the light, and infertility. Proteasome complexes missing RPN5a are less stable in vitro, suggesting that some of the rpn5a defects are caused by altered complex integrity. The rpn5a phenotype could be rescued by expression of either RPN5a or RPN5b, indicating functional redundancy. However, abnormal phenotypes generated by overexpression implied that paralog-specific functions also exist. Collectively, the data point to a specific role for RPN5 in the plant 26S proteasome and suggest that its two paralogous genes in Arabidopsis have both redundant and unique roles in development.
Figures







References
-
- Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., Nussaume, L., Noh, Y.S., Amasino, R., and Scheres, B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119 109–120. - PubMed
-
- Cary, A.J., Che, P., and Howell, S.H. (2002). Developmental events and shoot apical meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J. 32 867–877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

