Energetic determinants of protein binding specificity: insights into protein interaction networks
- PMID: 19253304
- PMCID: PMC7299235
- DOI: 10.1002/pmic.200800425
Energetic determinants of protein binding specificity: insights into protein interaction networks
Abstract
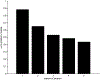
One of the challenges of the postgenomic era is to provide a more realistic representation of cellular processes by combining a systems biology description of functional networks with information on their interacting components. Here we carried out a systematic large-scale computational study on a structural protein-protein interaction network dataset in order to dissect thermodynamic characteristics of binding determining the interplay between protein affinity and specificity. As expected, interactions involving specific binding sites display higher affinities than those of promiscuous binding sites. Next, in order to investigate a possible role of modular distribution of hot spots in binding specificity, we divided binding sites into modules previously shown to be energetically independent. In general, hot spots that interact with different partners are located in different modules. We further observed that common hot spots tend to interact with partners exhibiting common binding motifs, whereas different hot spots tend to interact with partners with different motifs. Thus, energetic properties of binding sites provide insights into the way proteins modulate interactions with different partners. Knowledge of those factors playing a role in protein specificity is important for understanding how proteins acquire additional partners during evolution. It should also be useful in drug design.
Conflict of interest statement
The authors declare they have no conflict of interest.
Figures

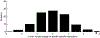
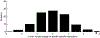
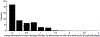
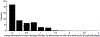
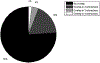
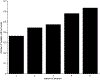

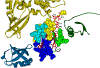
References
-
- Beckett D, Multilevel regulation of protein-protein interactions in biological circuitry. Phys Biol. 2005, 2, S67–73. - PubMed
-
- Kuriyan J, Eisenberg D, The origin of protein interactions and allostery in colocalization. Nature 2007, 450, 983–990. - PubMed
-
- Han JD, Bertin N, Hao T, Goldberg DS et al., Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 2004, 430, 88–93. - PubMed
-
- Kim PM, Lu LJ, Xia Y, Gerstein MB, Relating three-dimensional structures to protein networks provides evolutionary insights. Science 2006, 314, 1938–1941. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

