New bioinformatics approach to analyze gene expressions and signaling pathways reveals unique purine gene dysregulation profiles that distinguish between CD and UC
- PMID: 19253308
- PMCID: PMC2697273
- DOI: 10.1002/ibd.20893
New bioinformatics approach to analyze gene expressions and signaling pathways reveals unique purine gene dysregulation profiles that distinguish between CD and UC
Abstract
Background: Expression of purine genes is modulated by inflammation or experimental colitis and altered expression leads to disrupted gut function. We studied purine gene dysregulation profiles in inflammatory bowel disease (IBD) and determined whether they can distinguish between Crohn's disease (CD) and ulcerative colitis (UC) using Pathway Analysis and a new Comparative Analysis of Gene Expression and Selection (CAGES) method.
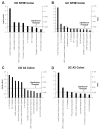
Methods: Raw datasets for 22 purine genes and 36 probe-sets from National Center for Biotechnology Information (NCBI) GEO (Gene Expression Omnibus) (http://www.ncbi.nlm.nih.gov/projects/geo/) were analyzed by National Cancer Institute (NCI) Biological Resources Branch (BRB) array tools for random-variance of multiple/36 t-tests in colonic mucosal biopsies or peripheral blood mononuclear cells (PBMCs) of CD, UC or control subjects. Dysregulation occurs in 59% of purine genes in IBD including ADORA3, CD73, ADORA2A, ADORA2B, ADAR, AMPD2, AMPD3, DPP4, P2RY5, P2RY6, P2RY13, P2RY14, and P2RX5.
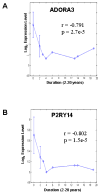
Results: In CD biopsies, expression of ADORA3, AMPD3, P2RY13, and P2RY5 were negatively correlated with acute inflammatory score, Crohn's Disease Activity Index (CDAI) or disease chronicity; P2RY14 was positively correlated in UC. In mucosal biopsies or PBMCs, CD and UC were distinguished by unique patterns of dysregulation (up- or downregulation) in purine genes. Purine gene dysregulation differs between PBMCs and biopsies and possibly between sexes for each disease. Ingenuity Pathway Analysis (IPA) revealed significant associations between alterations in the expression of CD73 (upregulation) or ADORA3 (downregulation) and inflammatory or purine genes (<or=10% of 57 genes) as well as G-protein coupled receptors, cAMP-dependent, and inflammatory pathways; IPA distinguishes CD from UC.
Conclusion: CAGES and Pathway Analysis provided novel evidence that UC and CD have distinct purine gene dysregulation signatures in association with inflammation, cAMP, or other signaling pathways. Disease-specific purine gene signature profiles and pathway associations may be of therapeutic, diagnostic, and functional relevance.
Figures

References
-
- Marak GE, Jr, de Kozak Y, Faure JP, et al. Pharmacologic modulation of acute ocular inflammation I Adenosine. Ophthalmic Res. 1988;20:220–226. - PubMed
-
- Schrier DJ, Lesch ME, Wright CD, et al. The anti-inflammatory effects of adenosine receptor agonists on the carrageenan-induced pleural inflammatory response in rats. J Immunol. 1990;145:1874–1879. - PubMed
-
- Hasko G, Szabo C. Regulation of cytokine and chemokine production by transmitters and co-transmitters of the autonomic nervous system. Biochem Pharmacol. 1998;56:1079–1087. - PubMed
-
- Hasko G, Kuhel DG, Chen JF, et al. Adenosine inhibits IL-12 and TNF-alpha production via adenosine A2a receptor-dependent and independent mechanisms. FASEB J. 2000;14:2065–2074. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
