Chemical issues addressing the construction of the distal Ni[cysteine-glycine-cysteine]2- site of acetyl CoA synthase: why not copper?
- PMID: 19253985
- PMCID: PMC2693717
- DOI: 10.1021/ic801628r
Chemical issues addressing the construction of the distal Ni[cysteine-glycine-cysteine]2- site of acetyl CoA synthase: why not copper?
Abstract
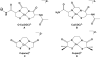
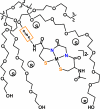
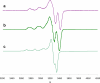
The discovery of the metallopeptide Ni(Cysteine-Glycine-Cysteine)(2-), Ni(CGC)(2-), in the A-cluster active site of Acetyl CoA Synthase has prompted the synthesis of many small molecule models which employ M(N(2)S(2)) complexes as metalloligands. In vitro studies have shown that nickel incorporates into the N(2)S(2) binding pocket even when copper is in the enzyme growth medium, while copper is preferentially taken up in the proximal site, displacing the catalytically active nickel. (Darnault, C.; Volbeda, A.; Kim, E.J.; Legrand, P.; Vernede, X.; Lindahl, P.A.; Fontecilla-Camps, J.C. Nat. Struct. Biol. 2003, 10, 271-279.) The work herein has been designed to address the chemical viability of copper(II) within the tripeptide N(2)S(2) ligand set. To this end, a series of CuN(2)S(2)(2-) complexes, the resin-bound, O-Cu(CGC)(2-) (A) and free Cu(CGC)(2-) (B) complexes, as well as Cu(ema)(2-) (C) and Cu(emi)(2-) (D) dianions, have been characterized by UV-vis, electron paramagnetic resonance (EPR), and electrospray ionization mass spectrometry (ESI-MS) spectroscopies, cyclic voltammetry (CV), and, where appropriate, X-ray diffraction studies, and compared to the Ni(II) congeners. EPR spectroscopic results have indicated that, in frozen N,N-dimethylformamide (DMF) solution, the copper complexes are distorted square planar structures with nitrogen and sulfur donors. This is consistent with X-ray diffraction measurements which also show copper(II) in a distorted square planar environment that is bereft of CuN(2)S(2)(2-) intermolecular interactions. Density-functional theory (DFT) calculations resulted in optimized structures that are consistent with crystallographic data and indicated highest occupied molecular orbital (HOMO)-singly occupied molecular orbital (SOMO) gaps of 5.01 and 4.68 eV for C and D, respectively. Optimized structures of Ni(ema)(2-) and Ni(emi)(2-) share the same basic characteristics as the copper(II) congeners. Electrochemical characterization of C and D resulted in a reversible Cu(III/II) couple at -1.20 V and - 1.40 V, respectively. Reactivity studies with Rh(CO)(2)(+) show similar donor capabilities for complexes A-D. Analysis of A shows that transmetalation does not occur. From competitive metal uptake studies on immobilized tripeptide it is concluded that the N(2)S(2)(4-) ligating unit has a slight preference for Cu(2+) over Ni(2+) and that the biosynthetic pathway responsible for constructing the distal site of ACS must be selective for nickel insertion or copper exclusion, or both.
Figures
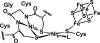
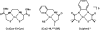



References
-
- Darnault C, Volbeda A, Kim EJ, Legrand P, Vernede X, Lindahl PA, Fontecilla-Camps JC. Nat. Struct. Biol. 2003;10:271–279. - PubMed
-
- Dokov IT, Iverson TM, Seravalli J, Ragsdale SW, Drennan CL. Science. 2002;298:567–572. - PubMed
-
- Bramlett MR, Tan X, Lindahl PA. J. Am. Chem. Soc. 2003;125:9316–9317. - PubMed
-
- Seravalli J, Xiao Y, Gu W, Cramer SP, Antholine WE, Krymov V, Gerfen GJ, Ragsdale SW. Biochemistry. 2004;43:3944–3955. - PubMed
-
- Nagashmia S, Nakasako M, Dohmae N, Tsujimura M, Takio K, Odaka M, Yohda M, Kamiya N, Endo I. Nat. Struct. Biol. 1998;5:347–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

