A novel view on stem cell development: analysing the shape of cellular genealogies
- PMID: 19254328
- PMCID: PMC6496517
- DOI: 10.1111/j.1365-2184.2009.00586.x
A novel view on stem cell development: analysing the shape of cellular genealogies
Abstract
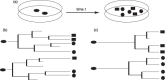
Objectives: The analysis of individual cell fates within a population of stem and progenitor cells is still a major experimental challenge in stem cell biology. However, new monitoring techniques, such as high-resolution time-lapse video microscopy, facilitate tracking and quantitative analysis of single cells and their progeny. Information on cellular development, divisional history and differentiation are naturally comprised into a pedigree-like structure, denoted as cellular genealogy. To extract reliable information concerning effecting variables and control mechanisms underlying cell fate decisions, it is necessary to analyse a large number of cellular genealogies.
Materials and methods: Here, we propose a set of statistical measures that are specifically tailored for the analysis of cellular genealogies. These measures address the degree and symmetry of cellular expansion, as well as occurrence and correlation of characteristic events such as cell death. Furthermore, we discuss two different methods for reconstruction of lineage fate decisions and show their impact on the interpretation of asymmetric developments. In order to illustrate these techniques, and to circumvent the present shortage of available experimental data, we obtain cellular genealogies from a single-cell-based mathematical model of haematopoietic stem cell organization.
Results and conclusions: Based on statistical analysis of cellular genealogies, we conclude that effects of external variables, such as growth conditions, are imprinted in their topology. Moreover, we demonstrate that it is essential to analyse timing of cell fate-specific changes and of occurrence of cell death events in the divisional context in order to understand the mechanisms of lineage commitment.
Figures






References
-
- Schroeder T (2005) Tracking hematopoiesis at the single cell level. Ann. N Y Acad. Sci. 1044, 201–209. - PubMed
-
- Roeder I, Lorenz R (2006) Asymmetry of stem cell fate and the potential impact of the niche: observations, simulations, and interpretations. Stem Cell Rev. 2, 171–180. - PubMed
-
- Morrison SJ, Shah NM, Anderson DJ (1997) Regulatory mechanisms in stem cell biology. Cell 88, 287–298. - PubMed
-
- Soneji S, Huang S, Loose M, Donaldson IJ, Patient R, Göttgens B, Enver T, May G (2007) Inference, validation, and dynamic modeling of transcription networks in multipotent hematopoietic cells. Ann. N Y Acad. Sci. 1106, 30–40. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

