A mesoscale model of DNA and its renaturation
- PMID: 19254530
- PMCID: PMC2717267
- DOI: 10.1016/j.bpj.2008.09.061
A mesoscale model of DNA and its renaturation
Abstract
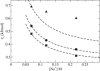
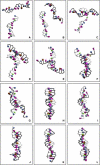
A mesoscale model of DNA is presented (3SPN.1), extending the scheme previously developed by our group. Each nucleotide is mapped onto three interaction sites. Solvent is accounted for implicitly through a medium-effective dielectric constant and electrostatic interactions are treated at the level of Debye-Hückel theory. The force field includes a weak, solvent-induced attraction, which helps mediate the renaturation of DNA. Model parameterization is accomplished through replica exchange molecular dynamics simulations of short oligonucleotide sequences over a range of composition and chain length. The model describes the melting temperature of DNA as a function of composition as well as ionic strength, and is consistent with heat capacity profiles from experiments. The dependence of persistence length on ionic strength is also captured by the force field. The proposed model is used to examine the renaturation of DNA. It is found that a typical renaturation event occurs through a nucleation step, whereby an interplay between repulsive electrostatic interactions and colloidal-like attractions allows the system to undergo a series of rearrangements before complete molecular reassociation occurs.
Figures








References
-
- Solis F.J., Olvera de la Cruz M. Attractive interactions between rodlike polyelectrolytes: polarization, crystallization, and packing. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1999;60:4496–4499. - PubMed
-
- Lyubartsev A.P., Nordenskiöld L. Monte Carlo simulation study of DNA polyelectrolyte properties in the presence of multivalent polyamine ions. J. Phys. Chem. B. 1997;101:4335–4342.
-
- Gil Montoro J.C., Abascal J.L.F. Ionic distribution around simple DNA models. I. Cylindrically averaged properties. J. Chem. Phys. 1995;103:8273–8284.
-
- Schlick T., Olson W.K. Supercoiled DNA energetics and dynamics by computer simulation. J. Mol. Biol. 1992;223:1089–1119. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

