Quantitative modeling and analysis of the transforming growth factor beta signaling pathway
- PMID: 19254534
- PMCID: PMC2717289
- DOI: 10.1016/j.bpj.2008.11.050
Quantitative modeling and analysis of the transforming growth factor beta signaling pathway
Abstract
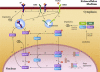
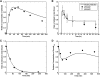
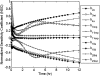
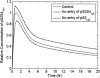
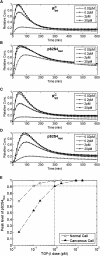
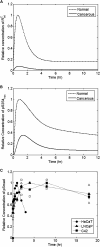
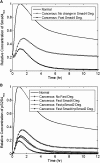
Transforming growth factor beta (TGF-beta) signaling, which regulates multiple cellular processes including proliferation, apoptosis, and differentiation, plays an important but incompletely understood role in normal and cancerous tissues. For instance, although TGF-beta functions as a tumor suppressor in the premalignant stages of tumorigenesis, paradoxically, it also seems to act as a tumor promoter in advanced cancer leading to metastasis. The mechanisms by which TGF-beta elicits such diverse responses during cancer progression are still not entirely clear. As a first step toward understanding TGF-beta signaling quantitatively, we have developed a comprehensive, dynamic model of the canonical TGF-beta pathway via Smad transcription factors. By describing how an extracellular signal of the TGF-beta ligand is sensed by receptors and transmitted into the nucleus through intracellular Smad proteins, the model provides quantitative insight into how TGF-beta-induced responses are modulated and regulated. Subsequent model analysis shows that mechanisms associated with Smad activation by ligand-activated receptor, nuclear complex formation among Smad proteins, and inactivation of ligand-activated Smad (e.g., degradation, dephosphorylation) may be critical for regulating TGF-beta-targeted functional responses. The model was also used to predict dynamic characteristics of the Smad-mediated pathway in abnormal cells, from which we generated four testable hypotheses regarding potential mechanisms by which TGF-beta's tumor-suppressive roles may appear to morph into tumor-promotion during cancer progression.
Figures



References
-
- Massague J. TGF-β signal transduction. Annu. Rev. Biochem. 1998;67:753–791. - PubMed
-
- Massague J., Gomis R.R. The logic of TGFβ signaling. FEBS Lett. 2006;580:2811–2820. - PubMed
-
- Shi Y., Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. - PubMed
-
- Massague J. How cells read TGF-β signals. Nat. Rev. Mol. Cell Biol. 2000;1:169–178. - PubMed
-
- Itoh S., Itoh F., Goumans M.J., Ten Dijke P. Signaling of transforming growth factor-β family members through Smad proteins. Eur. J. Biochem. 2000;267:6954–6967. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

