Selectivity and cooperativity of modulatory ions in a neurotransmitter receptor
- PMID: 19254535
- PMCID: PMC2717295
- DOI: 10.1016/j.bpj.2008.11.039
Selectivity and cooperativity of modulatory ions in a neurotransmitter receptor
Abstract
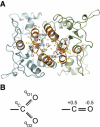
Ions play a modulatory role in many proteins. Kainate receptors, members of the ionotropic glutamate receptor family, require both monovalent anions and cations in the extracellular milieu for normal channel activity. Molecular dynamics simulations and extensive relative binding free energy calculations using thermodynamic integration were performed to elucidate the rank order of binding of monovalent cations, using x-ray crystal structures of the GluR5 kainate receptor dimers with bound cations from the alkali metal family. The simulations show good agreement with experiments and reveal that the underlying backbone structure of the binding site is one of the most rigid regions of the protein. A simplified model where the partial charge of coordinating oxygens was varied suggests that selectivity arises from the presence of two carboxylate groups. Furthermore, using a potential of mean force derived from umbrella sampling, we show that the presence of cations lower the energy barrier for anion approach and binding in the buried anion binding cavity.
Figures




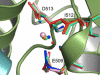
References
-
- Dingledine R., Borges K., Bowie D., Traynelis S.F. The glutamate receptor ion channels. Pharmacol. Rev. 1999;51:7–61. - PubMed
-
- Mayer M.L. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. - PubMed
-
- Hollmann M., Heinemann S.F. Cloned glutamate receptors. Annu. Rev. Neurosci. 1994;17:31–108. - PubMed
-
- Yamakura T., Shimoji K. Subunit and site specific pharmacology of the NMDA receptor channel. Prog. Neurobiol. 1999;59:279–298. - PubMed
-
- Lerma J., Morales M., Vicente M.A., Herreras O. Glutamate receptors of the kainate type and synaptic transmission. Trends Neurosci. 1997;20:9–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

