A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
- PMID: 19255093
- PMCID: PMC2673437
- DOI: 10.1093/nar/gkp092
A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Abstract
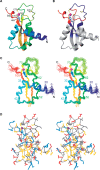
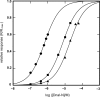
The helicase loader protein DnaI (the Bacillus subtilis homologue of Escherichia coli DnaC) is required to load the hexameric helicase DnaC (the B. subtilis homologue of E. coli DnaB) onto DNA at the start of replication. While the C-terminal domain of DnaI belongs to the structurally well-characterized AAA+ family of ATPases, the structure of the N-terminal domain, DnaI-N, has no homology to a known structure. Three-dimensional structure determination by nuclear magnetic resonance (NMR) spectroscopy shows that DnaI presents a novel fold containing a structurally important zinc ion. Surface plasmon resonance experiments indicate that DnaI-N is largely responsible for binding of DnaI to the hexameric helicase from B. stearothermophilus, which is a close homologue of the corresponding much less stable B. subtilis helicase.
Figures

Similar articles
-
Diverse Mechanisms of Helicase Loading during DNA Replication Initiation in Bacteria.J Bacteriol. 2023 Apr 25;205(4):e0048722. doi: 10.1128/jb.00487-22. Epub 2023 Mar 6. J Bacteriol. 2023. PMID: 36877032 Free PMC article. Review.
-
Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis.Nucleic Acids Res. 2006;34(18):5247-58. doi: 10.1093/nar/gkl690. Epub 2006 Sep 26. Nucleic Acids Res. 2006. PMID: 17003052 Free PMC article.
-
Molecular interplay between the replicative helicase DnaC and its loader protein DnaI from Geobacillus kaustophilus.J Mol Biol. 2009 Nov 13;393(5):1056-69. doi: 10.1016/j.jmb.2009.09.002. Epub 2009 Sep 8. J Mol Biol. 2009. PMID: 19744498
-
A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus.Nucleic Acids Res. 2002 Feb 15;30(4):966-74. doi: 10.1093/nar/30.4.966. Nucleic Acids Res. 2002. PMID: 11842108 Free PMC article.
-
Crystal structure of the complex of the interaction domains of Escherichia coli DnaB helicase and DnaC helicase loader: structural basis implying a distortion-accumulation mechanism for the DnaB ring opening caused by DnaC binding.J Biochem. 2020 Jan 1;167(1):1-14. doi: 10.1093/jb/mvz087. J Biochem. 2020. PMID: 31665315
Cited by
-
Physical Basis for the Loading of a Bacterial Replicative Helicase onto DNA.Mol Cell. 2019 Apr 4;74(1):173-184.e4. doi: 10.1016/j.molcel.2019.01.023. Epub 2019 Feb 20. Mol Cell. 2019. PMID: 30797687 Free PMC article.
-
Loading mechanisms of ring helicases at replication origins.Mol Microbiol. 2012 Apr;84(1):6-16. doi: 10.1111/j.1365-2958.2012.08012.x. Epub 2012 Mar 15. Mol Microbiol. 2012. PMID: 22417087 Free PMC article. Review.
-
Diverse Mechanisms of Helicase Loading during DNA Replication Initiation in Bacteria.J Bacteriol. 2023 Apr 25;205(4):e0048722. doi: 10.1128/jb.00487-22. Epub 2023 Mar 6. J Bacteriol. 2023. PMID: 36877032 Free PMC article. Review.
-
Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target.Curr Drug Targets. 2012 Mar;13(3):352-72. doi: 10.2174/138945012799424598. Curr Drug Targets. 2012. PMID: 22206257 Free PMC article. Review.
-
Structure of a helicase-helicase loader complex reveals insights into the mechanism of bacterial primosome assembly.Nat Commun. 2013;4:2495. doi: 10.1038/ncomms3495. Nat Commun. 2013. PMID: 24048025 Free PMC article.
References
-
- Schaeffer PM, Headlam MJ, Dixon NE. Protein-protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. - PubMed
-
- San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo J-M. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure. 1998;6:501–509. - PubMed
-
- Yuzhakov A, Turner J, O’Donnell M. Replisome assembly reveals the basis for asymmetric function in leading and lagging strand replication. Cell. 1996;86:877–886. - PubMed
-
- Davey MJ, O’Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr. Biol. 2003;13:R594–R596. - PubMed

