Methylation of H3 K4 and K79 is not strictly dependent on H2B K123 ubiquitylation
- PMID: 19255247
- PMCID: PMC2686411
- DOI: 10.1083/jcb.200812088
Methylation of H3 K4 and K79 is not strictly dependent on H2B K123 ubiquitylation
Abstract
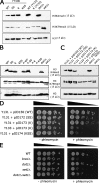
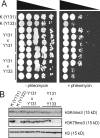
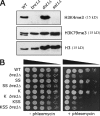
Covalent modifications of histone proteins have profound consequences on chromatin structure and function. Specific modification patterns constitute a code read by effector proteins. Studies from yeast found that H3 trimethylation at K4 and K79 is dependent on ubiquitylation of H2B K123, which is termed a "trans-tail pathway." In this study, we show that a strain unable to be ubiquitylated on H2B (K123R) is still proficient for H3 trimethylation at both K4 and K79, indicating that H3 methylation status is not solely dependent on H2B ubiquitylation. However, additional mutations in H2B result in loss of H3 methylation when combined with htb1-K123R. Consistent with this, we find that the original strain used to identify the trans-tail pathway has a genomic mutation that, when combined with H2B K123R, results in defective H3 methylation. Finally, we show that strains lacking the ubiquitin ligase Bre1 are defective for H3 methylation, suggesting that there is an additional Bre1 substrate that in combination with H2B K123 facilitates H3 methylation.
Figures


References
-
- Bostelman L.J., Keller A.M., Albrecht A.M., Arat A., Thompson J.S. 2007. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae.DNA Repair (Amst.). 6:383–395 - PubMed
-
- Briggs S.D., Xiao T., Sun Z.-W., Caldwell J.A., Shabanowitz J., Hunt D.F., Allis C.D., Strahl B.D. 2002. Gene silencing: trans-histone regulatory pathway in chromatin.Nature. 418:498. - PubMed
-
- Dehe P.M., Pamblanco M., Luciano P., Lebrun R., Moinier D., Sendra R., Verreault A., Tordera V., Geli V. 2005. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B.J. Mol. Biol. 353:477–484 - PubMed
-
- Dover J., Schneider J., Tawiah-Boateng M.A., Wood A., Dean K., Johnston M., Shilatifard A. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6.J. Biol. Chem. 277:28368–28371 - PubMed
-
- Downs J.A., Lowndes N.F., Jackson S.P. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair.Nature. 408:1001–1004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

