Identifying the proteins to which small-molecule probes and drugs bind in cells
- PMID: 19255428
- PMCID: PMC2649954
- DOI: 10.1073/pnas.0900191106
Identifying the proteins to which small-molecule probes and drugs bind in cells
Abstract
Most small-molecule probes and drugs alter cell circuitry by interacting with 1 or more proteins. A complete understanding of the interacting proteins and their associated protein complexes, whether the compounds are discovered by cell-based phenotypic or target-based screens, is extremely rare. Such a capability is expected to be highly illuminating--providing strong clues to the mechanisms used by small-molecules to achieve their recognized actions and suggesting potential unrecognized actions. We describe a powerful method combining quantitative proteomics (SILAC) with affinity enrichment to provide unbiased, robust and comprehensive identification of the proteins that bind to small-molecule probes and drugs. The method is scalable and general, requiring little optimization across different compound classes, and has already had a transformative effect on our studies of small-molecule probes. Here, we describe in full detail the application of the method to identify targets of kinase inhibitors and immunophilin binders.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


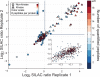
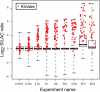

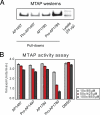
References
-
- Vesely J, et al. Inhibition of cyclin-dependent kinases by purine analogues. Eur J Biochem. 1994;224:771–786. - PubMed
-
- Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. - PubMed
-
- Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. - PubMed
-
- Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: Selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov. 2004;3:353–359. - PubMed
-
- Lindsay MA. Target discovery. Nat Rev Drug Discov. 2003;2:831–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

