Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy
- PMID: 19255432
- PMCID: PMC2649960
- DOI: 10.1073/pnas.0800442106
Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy
Abstract
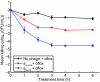
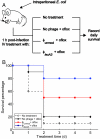
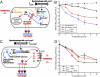
Antimicrobial drug development is increasingly lagging behind the evolution of antibiotic resistance, and as a result, there is a pressing need for new antibacterial therapies that can be readily designed and implemented. In this work, we engineered bacteriophage to overexpress proteins and attack gene networks that are not directly targeted by antibiotics. We show that suppressing the SOS network in Escherichia coli with engineered bacteriophage enhances killing by quinolones by several orders of magnitude in vitro and significantly increases survival of infected mice in vivo. In addition, we demonstrate that engineered bacteriophage can enhance the killing of antibiotic-resistant bacteria, persister cells, and biofilm cells, reduce the number of antibiotic-resistant bacteria that arise from an antibiotic-treated population, and act as a strong adjuvant for other bactericidal antibiotics (e.g., aminoglycosides and beta-lactams). Furthermore, we show that engineering bacteriophage to target non-SOS gene networks and to overexpress multiple factors also can produce effective antibiotic adjuvants. This work establishes a synthetic biology platform for the rapid translation and integration of identified targets into effective antibiotic adjuvants.
Conflict of interest statement
Conflict of interest statement: We have submitted a patent disclosure regarding the work described in this paper.
Figures

References
-
- Wise R. The relentless rise of resistance? J Antimicrob Chemother. 2004;54(2):306–310. - PubMed
-
- Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nature Reviews Microbiology. 2004;2(2):95–108. - PubMed
-
- Hall BG. Predicting the evolution of antibiotic resistance genes. Nature Reviews Microbiology. 2004;2(5):430–435. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305(5690):1622–1625. - PubMed
-
- Lewis K. Persister cells, dormancy and infectious disease. Nature Reviews Microbiology. 2007;5(1):48–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

