Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical, and fine structural analysis
- PMID: 19255771
- PMCID: PMC2761764
- DOI: 10.1007/s00418-009-0577-1
Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical, and fine structural analysis
Abstract
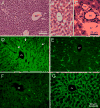
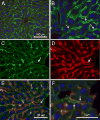
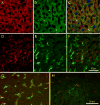
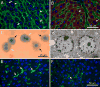
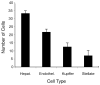
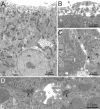
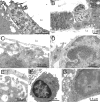
The cellular organization of normal mouse liver was studied using light and electron microscopy and quantitative immunocytochemical techniques. The general histological organization of the mouse liver is similar to livers of other mammalian species, with a lobular organization based on the distributions of portal areas and central venules. The parenchymal hepatocytes were detected with immunocytochemical techniques to recognize albumin or biotin containing cells. The macrophage Kupffer cells were identified with F4-80 immunocytochemistry, Ito stellate cells were identified with GFAP immunocytochemistry, and endothelial cells were labeled with the CD-34 antibody. Kupffer cells were labeled with intravascularly administered fluorescently labeled latex microspheres of both large (0.5 mum) and small (0.03 mum) diameters, while endothelial cells were labeled only with small diameter microspheres. Neither hepatocytes nor Ito stellate cells were labeled by intravascularly administered latex microspheres. The principal fine structural features of hepatocytes and non-parenchymal cells of mouse liver are similar to those reported for rat. Counts of immunocytochemically labeled cells with stained nuclei indicated that hepatocytes constituted approximately 52% of all labeled cells, Kupffer cells about 18%, Ito cells about 8%, and endothelial cells about 22% of all labeled cells. Approximately, 35% of the hepatocytes contained two nuclei; none of the Kupffer or Ito cells were double nucleated. The presence of canaliculi and a bile duct system appear similar to that reported for other species. The cellular organization of the mouse liver is quite similar to that of other mammalian species, confirming that the mouse presents a useful animal model for studies of liver structure and function.
Figures
References
-
- Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. - PubMed
-
- Aschoff L. Das Reticulo/endotheliale system. Ergebn Med Kinderheilk. 1924;26:1–118.
-
- Austyn JM, Gordon S. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur J Immunol. 1981;11:805–815. - PubMed
-
- Bartök I, Töth J, Remenar E, Viragh S. Fine structure of perisinusoidal cells in developing human and mouse liver. Acta Morphol Hung. 1983;31:337–352. - PubMed
-
- Bernuau D, Poliard A, Tournier I, Sala-Trepat J, Feldmann G. All hepatocytes are involved in the expression of the albumin gene in the normal adult rat: a demonstration by in situ hybridization and immunoperoxidase techniques. Cell Biol Int Rep. 1985;9:31–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

