Optimization of an unnatural base pair toward natural-like replication
- PMID: 19256568
- PMCID: PMC2901498
- DOI: 10.1021/ja807853m
Optimization of an unnatural base pair toward natural-like replication
Abstract
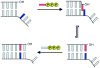
Predominantly hydrophobic unnatural nucleotides that selectively pair within duplex DNA as well as during polymerase-mediated replication have recently received much attention as the cornerstone of efforts to expand the genetic alphabet. We recently reported the results of a screen and subsequent lead hit optimization that led to identification of the unnatural base pair formed between the nucleotides dMMO2 and d5SICS. This unnatural base pair is replicated by the Klenow fragment of Escherichia coli DNA polymerase I with better efficiency and fidelity than other candidates reported in the literature. However, its replication remains significantly less efficient than a natural base pair, and further optimization is necessary for its practical use. To better understand and optimize the slowest step of replication of the unnatural base pair, the insertion of dMMO2 opposite d5SICS, we synthesized two dMMO2 derivatives, d5FM and dNaM, which differ from the parent nucleobase in terms of shape, hydrophobicity, and polarizability. We find that both derivatives are inserted opposite d5SICS more efficiently than dMMO2 and that overall the corresponding unnatural base pairs are generally replicated with higher efficiency and fidelity than the pair between dMMO2 and d5SICS. In fact, in the case of the dNaM:d5SICS heteropair, the efficiency of each individual step of replication approaches that of a natural base pair, and the minimum overall fidelity ranges from 10(3) to 10(4). In addition, the data allow us to propose a generalized model of unnatural base pair replication, which should aid in further optimization of the unnatural base pair and possibly in the design of additional unnatural base pairs that are replicated with truly natural-like efficiency and fidelity.
Figures
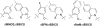
Similar articles
-
Major groove substituents and polymerase recognition of a class of predominantly hydrophobic unnatural base pairs.Chemistry. 2012 Jan 23;18(4):1231-9. doi: 10.1002/chem.201102066. Epub 2011 Dec 21. Chemistry. 2012. PMID: 22190386 Free PMC article.
-
Transcription of an expanded genetic alphabet.J Am Chem Soc. 2009 Apr 15;131(14):5046-7. doi: 10.1021/ja9006996. J Am Chem Soc. 2009. PMID: 19351201 Free PMC article.
-
Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet.J Am Chem Soc. 2008 Feb 20;130(7):2336-43. doi: 10.1021/ja078223d. Epub 2008 Jan 25. J Am Chem Soc. 2008. PMID: 18217762 Free PMC article.
-
Beyond A, C, G and T: augmenting nature's alphabet.Curr Opin Chem Biol. 2003 Dec;7(6):727-33. doi: 10.1016/j.cbpa.2003.10.011. Curr Opin Chem Biol. 2003. PMID: 14644182 Review.
-
The expanded genetic alphabet.Angew Chem Int Ed Engl. 2015 Oct 5;54(41):11930-44. doi: 10.1002/anie.201502890. Epub 2015 Aug 25. Angew Chem Int Ed Engl. 2015. PMID: 26304162 Free PMC article. Review.
Cited by
-
In Vivo Structure-Activity Relationships and Optimization of an Unnatural Base Pair for Replication in a Semi-Synthetic Organism.J Am Chem Soc. 2017 Aug 23;139(33):11427-11433. doi: 10.1021/jacs.7b03540. Epub 2017 Aug 10. J Am Chem Soc. 2017. PMID: 28796508 Free PMC article.
-
From polymerase engineering to semi-synthetic life: artificial expansion of the central dogma.RSC Chem Biol. 2022 Aug 9;3(10):1173-1197. doi: 10.1039/d2cb00116k. eCollection 2022 Oct 5. RSC Chem Biol. 2022. PMID: 36320892 Free PMC article. Review.
-
Major groove substituents and polymerase recognition of a class of predominantly hydrophobic unnatural base pairs.Chemistry. 2012 Jan 23;18(4):1231-9. doi: 10.1002/chem.201102066. Epub 2011 Dec 21. Chemistry. 2012. PMID: 22190386 Free PMC article.
-
Locating, tracing and sequencing multiple expanded genetic letters in complex DNA context via a bridge-base approach.Nucleic Acids Res. 2023 May 22;51(9):e52. doi: 10.1093/nar/gkad218. Nucleic Acids Res. 2023. PMID: 36971131 Free PMC article.
-
DNA damage and interstrand cross-link formation upon irradiation of aryl iodide C-nucleotide analogues.J Org Chem. 2010 Feb 5;75(3):535-44. doi: 10.1021/jo902071y. J Org Chem. 2010. PMID: 20067226 Free PMC article.
References
-
- Switzer C, Moroney SE, Benner SA. J Am Chem Soc. 1989;111:8322–8323.
-
- Geyer CR, Battersby TR, Benner SA. Structure (Camb) 2003;11:1485–1498. - PubMed
-
- Moser MJ, Marshall DJ, Greineri JK, Kieffer CD, Killeen AA, Ptacin JL, Richmond CS, Roesch EB, Scherrer CW, Sherrill CB, Van Hout CZ, Zanton SJ, Prudent JR. Clin Chem. 2003;49:407–414. - PubMed
-
- Prudent JR. Expert Rev Mol Diagn. 2006;6:245–252. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

