Kinetic folding studies of the P22 tailspike beta-helix domain reveal multiple unfolded states
- PMID: 19258192
- PMCID: PMC9295601
- DOI: 10.1016/j.bpc.2009.02.001
Kinetic folding studies of the P22 tailspike beta-helix domain reveal multiple unfolded states
Abstract
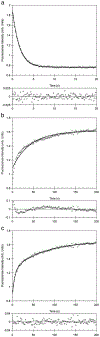
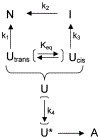
The beta-helix is an important protein fold in many pathogens, and is a challenging system for folding pathway prediction because it primarily is stabilized by non-local interactions along the primary sequence. A useful experimental model of this fold is a monomeric truncation of P22 tailspike protein, the beta-helix domain (bhx). This report describes a systematic in vitro study of the chemical denaturation and refolding of bhx. Results from equilibrium chemical denaturation experiments were consistent with a two-state folding mechanism, but showed only partial reversibility. Stopped-flow fluorescence studies showed a single unfolding step, but two refolding steps. The slow refolding step could be partly attributed to proline isomerization, based on an increased rate during refolding in the presence of PPIase and an increased relative amplitude of this step with increasing delay time in double-jump refolding experiments observed over delays of 5-100 s. However, double-jump refolding experiments with delay times longer than 100 s along with size exclusion chromatography and dynamic light scattering of refolding samples showed that the overall refolding yield decreased as bhx was unfolded for longer periods of time. Furthermore, the losses resulted from aggregate formation during refolding. This suggests that a change occurs over time in the unfolded or denatured state ensemble that increases the propensity for aggregation upon the shift to more native-favoring conditions. Alternatively aggregate nuclei may be able to form even under high denaturant conditions, and these subsequently grow and consume monomer when placed under native-favoring conditions.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

