The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts
- PMID: 19259985
- PMCID: PMC2918404
- DOI: 10.1002/jcb.22108
The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts
Abstract
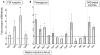
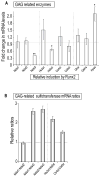
Heparan sulfate proteoglycans cooperate with basic fibroblast growth factor (bFGF/FGF2) signaling to control osteoblast growth and differentiation, as well as metabolic functions of osteoblasts. FGF2 signaling modulates the expression and activity of Runt-related transcription factor 2 (Runx2/Cbfa1), a key regulator of osteoblast proliferation and maturation. Here, we have characterized novel Runx2 target genes in osteoprogenitors under conditions that promote growth arrest while not yet permitting sustained phenotypic maturation. Runx2 enhances expression of genes related to proteoglycan-mediated signaling, including FGF receptors (e.g., FGFR2 and FGFR3) and proteoglycans (e.g., syndecans [Sdc1, Sdc2, Sdc3], glypicans [Gpc1], versican [Vcan]). Runx2 increases expression of the glycosyltransferase Exostosin-1 (Ext1) and heparanase, as well as alters the relative expression of N-linked sulfotransferases (Ndst1 = Ndst2 > Ndst3) and enzymes mediating O-linked sulfation of heparan sulfate (Hs2st > Hs6st) or chondroitin sulfate (Cs4st > Cs6st). Runx2 cooperates with FGF2 to induce expression of Sdc4 and the sulfatase Galns, but Runx2 and FGF2 suppress Gpc6, thus suggesting intricate Runx2 and FGF2 dependent changes in proteoglycan utilization. One functional consequence of Runx2 mediated modulations in proteoglycan-related gene expression is a change in the responsiveness of bone markers to FGF2 stimulation. Runx2 and FGF2 synergistically enhance osteopontin expression (>100 fold), while FGF2 blocks Runx2 induction of alkaline phosphatase. Our data suggest that Runx2 and the FGF/proteoglycan axis may form an extracellular matrix (ECM)-related regulatory feed-back loop that controls osteoblast proliferation and execution of the osteogenic program.
Figures




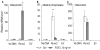

References
-
- Bae JS, Gutierrez S, Narla R, Pratap J, Devados R, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Javed A. Reconstitution of Runx2/Cbfa1-null cells identifies a requirement for BMP2 signaling through a Runx2 functional domain during osteoblast differentiation. J Cell Biochem. 2007;100:434–449. - PubMed
-
- Bodo M, Baroni T, Carinci F, Becchetti E, Bellucci C, Conte C, Pezzetti F, Evangelisti R, Tognon M, Carinci P. A regulatory role of fibroblast growth factor in the expression of decorin, biglycan, betaglycan and syndecan in osteoblasts from patients with Crouzon’s syndrome. Eur J Cell Biol. 1999;78:323–330. - PubMed
-
- Bonaventure J, El GV. Molecular and cellular bases of syndromic craniosynostoses. Expert Rev Mol Med. 2003;5:1–17. - PubMed
-
- Boudreaux JM, Towler DA. Synergistic induction of osteocalcin gene expression: identification of a bipartite element conferring fibroblast growth factor 2 and cyclic AMP responsiveness in the rat osteocalcin promoter. J Biol Chem. 1996;271:7508–7515. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

