A tale of tails: insights into the coordination of 3' end processing during homologous recombination
- PMID: 19260026
- PMCID: PMC2958051
- DOI: 10.1002/bies.200800195
A tale of tails: insights into the coordination of 3' end processing during homologous recombination
Abstract
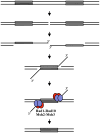
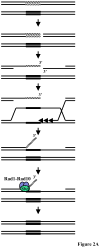
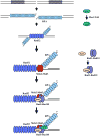
Eukaryotic genomes harbor a large number of homologous repeat sequences that are capable of recombining. Their potential to disrupt genome stability highlights the need to understand how homologous recombination processes are coordinated. The Saccharomyces cerevisiae Rad1-Rad10 endonuclease performs an essential role in recombination between repeated sequences, by processing 3' single-stranded intermediates formed during single-strand annealing and gene conversion events. Several recent studies have focused on factors involved in Rad1-Rad10-dependent removal of 3' nonhomologous tails during homologous recombination, including Msh2-Msh3, Slx4, and the newly identified Saw1 protein. Together, this new work provides a model for how Rad1-Rad10-dependent end processing is coordinated: Msh2-Msh3 stabilizes and prepares double-strand/single-strand junctions for Rad1-Rad10 cleavage, Saw1 recruits Rad1-Rad10 to 3' tails, and Slx4 mediates crosstalk between the DNA damage checkpoint machinery and Rad1-Rad10.
Figures

Similar articles
-
Coordination of Rad1-Rad10 interactions with Msh2-Msh3, Saw1 and RPA is essential for functional 3' non-homologous tail removal.Nucleic Acids Res. 2018 Jun 1;46(10):5075-5096. doi: 10.1093/nar/gky254. Nucleic Acids Res. 2018. PMID: 29660012 Free PMC article.
-
Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates.Mol Cell. 2008 May 9;30(3):325-35. doi: 10.1016/j.molcel.2008.02.028. Mol Cell. 2008. PMID: 18471978 Free PMC article.
-
Role of Saw1 in Rad1/Rad10 complex assembly at recombination intermediates in budding yeast.EMBO J. 2013 Feb 6;32(3):461-72. doi: 10.1038/emboj.2012.345. Epub 2013 Jan 8. EMBO J. 2013. PMID: 23299942 Free PMC article.
-
Bacterial DNA repair genes and their eukaryotic homologues: 5. The role of recombination in DNA repair and genome stability.Acta Biochim Pol. 2007;54(3):483-94. Epub 2007 Sep 23. Acta Biochim Pol. 2007. PMID: 17893749 Review.
-
Double-strand break repair and homologous recombination in Schizosaccharomyces pombe.Yeast. 2006 Oct 15;23(13):963-76. doi: 10.1002/yea.1414. Yeast. 2006. PMID: 17072889 Review.
Cited by
-
Rad1 and Rad10 Tied to Photolyase Regulators Protect Insecticidal Fungal Cells from Solar UV Damage by Photoreactivation.J Fungi (Basel). 2022 Oct 25;8(11):1124. doi: 10.3390/jof8111124. J Fungi (Basel). 2022. PMID: 36354891 Free PMC article.
-
Sumoylation of the Rad1 nuclease promotes DNA repair and regulates its DNA association.Nucleic Acids Res. 2014 Jun;42(10):6393-404. doi: 10.1093/nar/gku300. Epub 2014 Apr 20. Nucleic Acids Res. 2014. PMID: 24753409 Free PMC article.
-
Removal of reactive oxygen species-induced 3'-blocked ends by XPF-ERCC1.Chem Res Toxicol. 2011 Nov 21;24(11):1876-81. doi: 10.1021/tx200221j. Epub 2011 Oct 18. Chem Res Toxicol. 2011. PMID: 22007867 Free PMC article.
-
Multi-Faceted Roles of ERCC1-XPF Nuclease in Processing Non-B DNA Structures.DNA (Basel). 2022 Dec;2(4):231-247. doi: 10.3390/dna2040017. Epub 2022 Oct 11. DNA (Basel). 2022. PMID: 40766048 Free PMC article.
-
Novel insights into the ecDNA formation mechanism involving MSH3 in methotrexate‑resistant human colorectal cancer cells.Int J Oncol. 2023 Dec;63(6):134. doi: 10.3892/ijo.2023.5582. Epub 2023 Oct 27. Int J Oncol. 2023. PMID: 37888748 Free PMC article.
References
-
- Surtees JA, Alani E. Mismatch Repair Factor Msh2-Msh3 Binds and Alters the Conformation of Branched DNA Structures Predicted to Form During Genetic Recombination. J Mol Biol. 2006;360:523–526. - PubMed
-
- Li W-H, Gu Z, Wang H, Nekrutenko A. Evolutionary Analyses of the Human Genome. Nature. 2001;409:847–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

