A tale of tails: insights into the coordination of 3' end processing during homologous recombination
- PMID: 19260026
- PMCID: PMC2958051
- DOI: 10.1002/bies.200800195
A tale of tails: insights into the coordination of 3' end processing during homologous recombination
Abstract
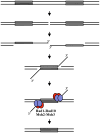
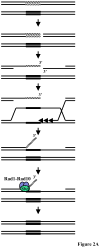
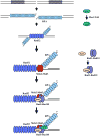
Eukaryotic genomes harbor a large number of homologous repeat sequences that are capable of recombining. Their potential to disrupt genome stability highlights the need to understand how homologous recombination processes are coordinated. The Saccharomyces cerevisiae Rad1-Rad10 endonuclease performs an essential role in recombination between repeated sequences, by processing 3' single-stranded intermediates formed during single-strand annealing and gene conversion events. Several recent studies have focused on factors involved in Rad1-Rad10-dependent removal of 3' nonhomologous tails during homologous recombination, including Msh2-Msh3, Slx4, and the newly identified Saw1 protein. Together, this new work provides a model for how Rad1-Rad10-dependent end processing is coordinated: Msh2-Msh3 stabilizes and prepares double-strand/single-strand junctions for Rad1-Rad10 cleavage, Saw1 recruits Rad1-Rad10 to 3' tails, and Slx4 mediates crosstalk between the DNA damage checkpoint machinery and Rad1-Rad10.
Figures

References
-
- Surtees JA, Alani E. Mismatch Repair Factor Msh2-Msh3 Binds and Alters the Conformation of Branched DNA Structures Predicted to Form During Genetic Recombination. J Mol Biol. 2006;360:523–526. - PubMed
-
- Li W-H, Gu Z, Wang H, Nekrutenko A. Evolutionary Analyses of the Human Genome. Nature. 2001;409:847–849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

