Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase
- PMID: 19260676
- PMCID: PMC2756614
- DOI: 10.1021/bi900039e
Structural and thermodynamic analysis of a conformationally strained circular permutant of barnase
Abstract
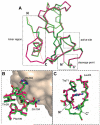
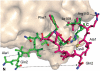
Circular permutation of a protein covalently links its original termini and creates new ends at another location. To maintain the stability of the permuted structure, the termini are typically bridged by a peptide long enough to span the original distance between them. Here, we take the opposite approach and employ a very short linker to introduce conformational strain into a protein by forcing its termini together. We join the N- and C-termini of the small ribonuclease barnase (normally 27.2 A distant) with a single Cys residue and introduce new termini at a surface loop, to create pBn. Compared to a similar variant permuted with an 18-residue linker, permutation with a single amino acid dramatically destabilizes barnase. Surprisingly, pBn is folded at 10 degrees C and possesses near wild-type ribonuclease activity. The 2.25 A X-ray crystal structure of pBn reveals how the barnase fold is able to adapt to permutation, partially defuse conformational strain, and preserve enzymatic function. We demonstrate that strain in pBn can be relieved by cleaving the linker with a chemical reagent. Catalytic activity of both uncleaved (strained) pBn and cleaved (relaxed) pBn is proportional to their thermodynamic stabilities, i.e., the fraction of folded molecules. The stability and activity of cleaved pBn are dependent on protein concentration. At concentrations above approximately 2 microM, cleaving pBn is predicted to increase the fraction of folded molecules and thus enhance ribonuclease activity at 37 degrees C. This study suggests that introducing conformational strain by permutation, and releasing strain by cleavage, is a potential mechanism for engineering an artificial zymogen.
Figures






References
-
- Perez-Jiminez R, Garcia-Manyes S, Ainavarapu SRK, Fernandez JM. Mechanical unfolding pathways of the enhanced yellow fluorescent protein revealed by single molecule force spectroscopy. J. Biol. Chem. 2006;281:40010–40014. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

