Polypurine hairpins directed against the template strand of DNA knock down the expression of mammalian genes
- PMID: 19261618
- PMCID: PMC2670163
- DOI: 10.1074/jbc.M900981200
Polypurine hairpins directed against the template strand of DNA knock down the expression of mammalian genes
Abstract
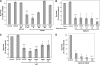
We analyzed whether polypurine hairpins (PPRHs) had the ability to knock down gene expression. These hairpins are formed by two antiparallel purine domains linked by a loop that allows the formation of Hoogsteen bonds between both domains and Watson-Crick bonds with the target polypyrimidine sequence, forming triplex structures. To set up the experimental conditions, the human dhfr gene was used as a model. The PPRHs were designed toward the template strand of DNA. The transfection of the human breast cancer cell line SKBR3 with these template hairpins against the dhfr gene produced higher than 90% of cell mortality. Template PPRHs produced a decrease in DHFR mRNA, protein, and its corresponding enzymatic activity. In addition, the activity of DHFR PPRHs was tested against breast cancer cells resistant to methotrexate, observing high cell mortality. Given the difficulty in finding long polypyrimidine stretches, we studied how to compensate for the presence of purine interruptions in the polypyrimidine target sequence. The stability of PPRH was measured, resulting in a surprisingly long half-life of about 5 days. Finally, to test the generality of usage, template PPRHs were employed against two important genes involved in cell proliferation, telomerase and survivin, producing 80 and 95% of cell death, respectively. Taken together our results show the ability of antiparallel purine hairpins to bind the template strand of double strand DNA and to decrease gene transcription. Thus, PPRHs can be considered as a new type of molecules to modulate gene expression.
Figures



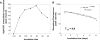



Similar articles
-
Coding polypurine hairpins cause target-induced cell death in breast cancer cells.Hum Gene Ther. 2011 Apr;22(4):451-63. doi: 10.1089/hum.2010.102. Epub 2011 Feb 16. Hum Gene Ther. 2011. PMID: 20942657
-
Polypurine reverse Hoogsteen hairpins as a gene therapy tool against survivin in human prostate cancer PC3 cells in vitro and in vivo.Biochem Pharmacol. 2013 Dec 1;86(11):1541-54. doi: 10.1016/j.bcp.2013.09.013. Epub 2013 Sep 23. Biochem Pharmacol. 2013. PMID: 24070653
-
Polypurine Reverse Hoogsteen Hairpins as a Gene Silencing Tool for Cancer.Curr Med Chem. 2017;24(26):2809-2826. doi: 10.2174/0929867324666170301114127. Curr Med Chem. 2017. PMID: 28260512 Review.
-
Effect of Polypurine Reverse Hoogsteen Hairpins on Relevant Cancer Target Genes in Different Human Cell Lines.Nucleic Acid Ther. 2015 Aug;25(4):198-208. doi: 10.1089/nat.2015.0531. Epub 2015 Jun 4. Nucleic Acid Ther. 2015. PMID: 26042602
-
Nucleic acids therapeutics using PolyPurine Reverse Hoogsteen hairpins.Biochem Pharmacol. 2021 Jul;189:114371. doi: 10.1016/j.bcp.2020.114371. Epub 2020 Dec 16. Biochem Pharmacol. 2021. PMID: 33338475 Review.
Cited by
-
Targeting KRAS Regulation with PolyPurine Reverse Hoogsteen Oligonucleotides.Int J Mol Sci. 2022 Feb 14;23(4):2097. doi: 10.3390/ijms23042097. Int J Mol Sci. 2022. PMID: 35216221 Free PMC article.
-
Polypurine reverse-Hoogsteen (PPRH) oligonucleotides can form triplexes with their target sequences even under conditions where they fold into G-quadruplexes.Sci Rep. 2017 Jan 9;7:39898. doi: 10.1038/srep39898. Sci Rep. 2017. PMID: 28067256 Free PMC article.
-
In Vitro and In Vivo Effects of the Combination of Polypurine Reverse Hoogsteen Hairpins against HER-2 and Trastuzumab in Breast Cancer Cells.Int J Mol Sci. 2023 Apr 11;24(8):7073. doi: 10.3390/ijms24087073. Int J Mol Sci. 2023. PMID: 37108234 Free PMC article.
-
Repair of single-point mutations by polypurine reverse Hoogsteen hairpins.Hum Gene Ther Methods. 2014 Oct;25(5):288-302. doi: 10.1089/hgtb.2014.049. Epub 2014 Oct 14. Hum Gene Ther Methods. 2014. PMID: 25222154 Free PMC article.
-
Gene Correction of Point Mutations Using PolyPurine Reverse Hoogsteen Hairpins Technology.Front Genome Ed. 2020 Oct 29;2:583577. doi: 10.3389/fgeed.2020.583577. eCollection 2020. Front Genome Ed. 2020. PMID: 34713221 Free PMC article. Review.
References
-
- Chan, P. P., and Glazer, P. M. (1997) J. Mol. Med. 75, 267-282 - PubMed
-
- Giovannangeli, C., and Hâeláene, C. (1997) Antisense Nucleic Acid Drug Dev. 7, 413-421 - PubMed
-
- Casey, B. P., and Glazer, P. M. (2001) Prog. Nucleic Acids Res. Mol. Biol. 67, 163-192 - PubMed
-
- Felsenfeld, G., and Rich, A. (1957) Biochim. Biophys. Acta 26, 457-468 - PubMed
-
- Ryan, K., and Kool, E. T. (1998) Chem. Biol. 5, 59-67 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

