A modified hyperplane clustering algorithm allows for efficient and accurate clustering of extremely large datasets
- PMID: 19261720
- PMCID: PMC2672630
- DOI: 10.1093/bioinformatics/btp123
A modified hyperplane clustering algorithm allows for efficient and accurate clustering of extremely large datasets
Abstract
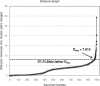
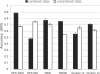
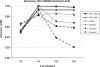
Motivation: As the number of publically available microarray experiments increases, the ability to analyze extremely large datasets across multiple experiments becomes critical. There is a requirement to develop algorithms which are fast and can cluster extremely large datasets without affecting the cluster quality. Clustering is an unsupervised exploratory technique applied to microarray data to find similar data structures or expression patterns. Because of the high input/output costs involved and large distance matrices calculated, most of the algomerative clustering algorithms fail on large datasets (30,000 + genes/200 + arrays). In this article, we propose a new two-stage algorithm which partitions the high-dimensional space associated with microarray data using hyperplanes. The first stage is based on the Balanced Iterative Reducing and Clustering using Hierarchies algorithm with the second stage being a conventional k-means clustering technique. This algorithm has been implemented in a software tool (HPCluster) designed to cluster gene expression data. We compared the clustering results using the two-stage hyperplane algorithm with the conventional k-means algorithm from other available programs. Because, the first stage traverses the data in a single scan, the performance and speed increases substantially. The data reduction accomplished in the first stage of the algorithm reduces the memory requirements allowing us to cluster 44,460 genes without failure and significantly decreases the time to complete when compared with popular k-means programs. The software was written in C# (.NET 1.1).
Availability: The program is freely available and can be downloaded from http://www.amdcc.org/bioinformatics/bioinformatics.aspx.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

Similar articles
-
FLAME, a novel fuzzy clustering method for the analysis of DNA microarray data.BMC Bioinformatics. 2007 Jan 4;8:3. doi: 10.1186/1471-2105-8-3. BMC Bioinformatics. 2007. PMID: 17204155 Free PMC article.
-
Discovering biclusters in gene expression data based on high-dimensional linear geometries.BMC Bioinformatics. 2008 Apr 23;9:209. doi: 10.1186/1471-2105-9-209. BMC Bioinformatics. 2008. PMID: 18433477 Free PMC article.
-
Detecting clusters of different geometrical shapes in microarray gene expression data.Bioinformatics. 2005 May 1;21(9):1927-34. doi: 10.1093/bioinformatics/bti251. Epub 2005 Jan 12. Bioinformatics. 2005. PMID: 15647300
-
Computational cluster validation in post-genomic data analysis.Bioinformatics. 2005 Aug 1;21(15):3201-12. doi: 10.1093/bioinformatics/bti517. Epub 2005 May 24. Bioinformatics. 2005. PMID: 15914541 Review.
-
Matrix factorisation methods applied in microarray data analysis.Int J Data Min Bioinform. 2010;4(1):72-90. doi: 10.1504/ijdmb.2010.030968. Int J Data Min Bioinform. 2010. PMID: 20376923 Free PMC article. Review.
Cited by
-
Mycophenolic acid inhibits migration and invasion of gastric cancer cells via multiple molecular pathways.PLoS One. 2013 Nov 15;8(11):e81702. doi: 10.1371/journal.pone.0081702. eCollection 2013. PLoS One. 2013. PMID: 24260584 Free PMC article.
-
Cell Cycle M-Phase Genes Are Highly Upregulated in Anaplastic Thyroid Carcinoma.Thyroid. 2017 Feb;27(2):236-252. doi: 10.1089/thy.2016.0285. Epub 2016 Dec 15. Thyroid. 2017. PMID: 27796151 Free PMC article.
-
CLIC: clustering analysis of large microarray datasets with individual dimension-based clustering.Nucleic Acids Res. 2010 Jul;38(Web Server issue):W246-53. doi: 10.1093/nar/gkq516. Epub 2010 Jun 6. Nucleic Acids Res. 2010. PMID: 20529873 Free PMC article.
-
Celda: a Bayesian model to perform co-clustering of genes into modules and cells into subpopulations using single-cell RNA-seq data.NAR Genom Bioinform. 2022 Sep 13;4(3):lqac066. doi: 10.1093/nargab/lqac066. eCollection 2022 Sep. NAR Genom Bioinform. 2022. PMID: 36110899 Free PMC article.
-
Clustering of High Throughput Gene Expression Data.Comput Oper Res. 2012 Dec;39(12):3046-3061. doi: 10.1016/j.cor.2012.03.008. Comput Oper Res. 2012. PMID: 23144527 Free PMC article.
References
-
- Chen G, et al. Evaluation and comparison of clustering algorithms in anglyzing ES cell gene expression data. Stat. Sin. 2002;12:241–262.
-
- Dash M, et al. Fast hierarchical clustering and its validation. Data Knowl. Eng. 2003;44:109–138.
-
- Datta S, Datta S. Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics. 2003;19:459–466. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

