Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing
- PMID: 19261920
- PMCID: PMC2671620
- DOI: 10.1093/jxb/erp038
Functional analysis of the isoforms of an ABI3-like factor of Pisum sativum generated by alternative splicing
Abstract
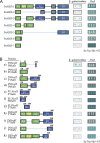
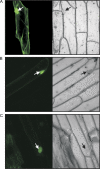
At least seven isoforms (PsABI3-1 to PsABI3-7) of a putative, pea ABI3-like factor, originated by alternative splicing, have been identified after cDNA cloning. A similar variability had previously only been described for monocot genes. The full-length isoform, PsABI3-1, contains the typical N-terminal acidic domains and C-terminal basic subdomains, B1 to B3. Reverse transcriptase-PCR analysis revealed that the gene is expressed just in seeds, starting at middle embryogenesis; no gene products are observed in embryo axes after 18 h post-imbibition although they are more persistent in cotyledons. The activity of the isoforms was studied by yeast one-hybrid assays. When yeast was transformed with the isoforms fused to the DNA binding domain of Gal4p, only the polypeptides PsABI3-2 and PsABI3-7 failed to complement the activity of Gal4p. Acidic domains A1 and A2 exhibit transactivating activity, but the former requires a small C-terminal extension to be active. Yeast two-hybrid analysis showed that PsABI3 is able to heterodimerize with Arabidopsis thaliana ABI5, thus proving that PsABI3 is functionally active. The minimum requirement for the interaction PsABI3-AtABI5 is the presence of the subdomain B1 with an extension, 81 amino acids long, at their C-terminal side. Finally, a transient onion transformation assay showed that both the active PsABI3-1 and the inactive PsABI3-2 isoforms are localized to nuclei. Considering that the major isoforms remain approximately constant in developing seeds although their relative proportion varied, the possible role of splicing in the regulatory network of ABA signalling is discussed.
Figures




Similar articles
-
A novel lipid transfer protein from the pea Pisum sativum: isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties.BMC Plant Biol. 2016 Apr 30;16:107. doi: 10.1186/s12870-016-0792-6. BMC Plant Biol. 2016. PMID: 27137920 Free PMC article.
-
Cloning and tissue-specific expression of predicted Pisum sativum actin isoform PEAc14-1.Biochem Genet. 2013 Oct;51(9-10):722-7. doi: 10.1007/s10528-013-9601-1. Epub 2013 May 28. Biochem Genet. 2013. PMID: 23712761 No abstract available.
-
Differential expression and functional analysis of three calmodulin isoforms in germinating pea (Pisum sativum L.) seeds.Plant J. 2002 Nov;32(4):481-93. doi: 10.1046/j.1365-313x.2002.01409.x. Plant J. 2002. PMID: 12445120
-
Arabidopsis genes encoding components of the chloroplastic protein import apparatus.Plant Physiol. 2001 Apr;125(4):1567-76. doi: 10.1104/pp.125.4.1567. Plant Physiol. 2001. PMID: 11299338 Free PMC article. Review.
-
Roles and regulatory patterns of protein isoforms in plant adaptation and development.New Phytol. 2025 Mar;245(5):1887-1896. doi: 10.1111/nph.20327. Epub 2024 Dec 7. New Phytol. 2025. PMID: 39645578 Review.
Cited by
-
Molecular Mechanisms Underlying Abscisic Acid/Gibberellin Balance in the Control of Seed Dormancy and Germination in Cereals.Front Plant Sci. 2018 May 23;9:668. doi: 10.3389/fpls.2018.00668. eCollection 2018. Front Plant Sci. 2018. PMID: 29875780 Free PMC article. Review.
-
Functional characterization of two alternatively spliced transcripts of tomato ABSCISIC ACID INSENSITIVE3 (ABI3) gene.Plant Mol Biol. 2013 May;82(1-2):131-45. doi: 10.1007/s11103-013-0044-1. Epub 2013 Mar 17. Plant Mol Biol. 2013. PMID: 23504452
-
Transcriptome Analysis of Alternative Splicing Events Induced by Arbuscular Mycorrhizal Fungi (Rhizophagus irregularis) in Pea (Pisum sativum L.) Roots.Plants (Basel). 2020 Dec 3;9(12):1700. doi: 10.3390/plants9121700. Plants (Basel). 2020. PMID: 33287282 Free PMC article.
-
Alternative Splicing as a Regulator of Early Plant Development.Front Plant Sci. 2018 Aug 15;9:1174. doi: 10.3389/fpls.2018.01174. eCollection 2018. Front Plant Sci. 2018. PMID: 30158945 Free PMC article. Review.
-
Aethionema arabicum genome annotation using PacBio full-length transcripts provides a valuable resource for seed dormancy and Brassicaceae evolution research.Plant J. 2021 Apr;106(1):275-293. doi: 10.1111/tpj.15161. Epub 2021 Feb 8. Plant J. 2021. PMID: 33453123 Free PMC article.
References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Deidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York: John Wiley and Sons; 1995.
-
- Bobb AJ, Eiben HG, Bustos MM. PvAlf, an embryo-specific acidic transcriptional activator enhances gene expression from phaseolin and phytohemagglutinin promoters. The Plant Journal. 1995;8:331–343. - PubMed
-
- Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425–435. - PubMed
-
- Castillo J, Genovés A, Franco L, Rodrigo MI. A multifunctional bicupin serves as precursor for a chromosomal protein of Pisum sativum seeds. Journal of Experimental Botany. 2005;56:3159–3169. - PubMed
-
- Castillo J, Rodrigo MI, Márquez JA, Zúñiga A, Franco L. A pea nuclear protein that is induced by dehydration belongs to the vicilin superfamily. European Journal of Biochemistry. 2000;267:2156–2165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

