The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae
- PMID: 19262693
- PMCID: PMC2650407
- DOI: 10.1371/journal.pone.0004670
The number and transmission of [PSI] prion seeds (Propagons) in the yeast Saccharomyces cerevisiae
Abstract
Background: Yeast (Saccharomyces cerevisiae) prions are efficiently propagated and the on-going generation and transmission of prion seeds (propagons) to daughter cells during cell division ensures a high degree of mitotic stability. The reversible inhibition of the molecular chaperone Hsp104p by guanidine hydrochloride (GdnHCl) results in cell division-dependent elimination of yeast prions due to a block in propagon generation and the subsequent dilution out of propagons by cell division.
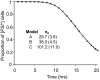
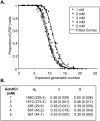
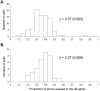
Principal findings: Analysing the kinetics of the GdnHCl-induced elimination of the yeast [PSI+] prion has allowed us to develop novel statistical models that aid our understanding of prion propagation in yeast cells. Here we describe the application of a new stochastic model that allows us to estimate more accurately the mean number of propagons in a [PSI+] cell. To achieve this accuracy we also experimentally determine key cell reproduction parameters and show that the presence of the [PSI+] prion has no impact on these key processes. Additionally, we experimentally determine the proportion of propagons transmitted to a daughter cell and show this reflects the relative cell volume of mother and daughter cells at cell division.
Conclusions: While propagon generation is an ATP-driven process, the partition of propagons to daughter cells occurs by passive transfer via the distribution of cytoplasm. Furthermore, our new estimates of n(0), the number of propagons per cell (500-1000), are some five times higher than our previous estimates and this has important implications for our understanding of the inheritance of the [PSI+] and the spontaneous formation of prion-free cells.
Conflict of interest statement
Figures

References
-
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. - PubMed
-
- Benkemoun L, Saupe SJ. Prion proteins as genetic material in fungi. Fungal Genet Biol. 2006;43:789–803. - PubMed
-
- Cox BS. [PSI], a cytoplasmic suppressor of super-suppressors in yeast. Heredity. 1965;20:505–521.
-
- Tuite MF, Cox BS. The [PSI +] prion of yeast: a problem of inheritance. Methods. 2006;39:9–22. - PubMed
-
- Tuite MF, Byrne LJ, Josse L, Ness F, Koloteva-Levine N, et al. Yeast prions and their analysis in vivo. In: Stansfield I, Stark MJR, editors. Yeast Gene Analysis: Academic Press; 2007. pp. 491–525.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

