c-Myc and Rel/NF-kappaB are the two master transcriptional systems activated in the latency III program of Epstein-Barr virus-immortalized B cells
- PMID: 19264782
- PMCID: PMC2682111
- DOI: 10.1128/JVI.02264-08
c-Myc and Rel/NF-kappaB are the two master transcriptional systems activated in the latency III program of Epstein-Barr virus-immortalized B cells
Abstract
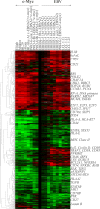
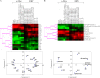
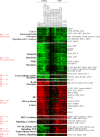
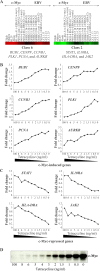
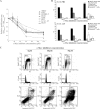
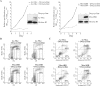
The Epstein-Barr virus (EBV) latency III program imposed by EBNA2 and LMP1 is directly responsible for immortalization of B cells in vitro and is thought to mediate most immunodeficiency-related posttransplant lymphoproliferative diseases in vivo. To answer the question whether and how this proliferation program is related to c-Myc, we have established the transcriptome of both c-Myc and EBV latency III proliferation programs using a Lymphochip specialized microarray. In addition to EBV-positive latency I Burkitt lymphoma lines and lymphoblastoid cell lines (LCLs), we used an LCL expressing an estrogen-regulatable EBNA2 fusion protein (EREB2-5) and derivative B-cell lines expressing a constitutively active or tetracycline-regulatable c-myc gene. A total of 897 genes were found to be fourfold or more up- or downregulated in either one or both proliferation programs compared to the expression profile of resting EREB2-5 cells. A total of 661 (74%) of these were regulated similarly in both programs. Numerous repressed genes were known targets of STAT1, and most induced genes were known to be upregulated by c-Myc and to be involved in cell proliferation. In keeping with the gene expression patterns, inactivation of c-Myc by a chemical inhibitor or by conditional expression of dominant-negative c-Myc and Max mutants led to proliferation arrest of LCLs. Most genes differently regulated in both proliferation programs corresponded to genes induced by NF-kappaB in LCLs, and many of them coded for immunoregulatory and/or antiapoptotic molecules. Thus, c-Myc and NF-kappaB are the two main transcription factors responsible for the phenotype, growth pattern, and biological properties of cells driven into proliferation by EBV.
Figures
Similar articles
-
EBV latency III immortalization program sensitizes B cells to induction of CD95-mediated apoptosis via LMP1: role of NF-kappaB, STAT1, and p53.Blood. 2006 Mar 1;107(5):2070-8. doi: 10.1182/blood-2005-05-2053. Epub 2005 Nov 29. Blood. 2006. PMID: 16317104
-
RNA Sequencing Analyses of Gene Expression during Epstein-Barr Virus Infection of Primary B Lymphocytes.J Virol. 2019 Jun 14;93(13):e00226-19. doi: 10.1128/JVI.00226-19. Print 2019 Jul 1. J Virol. 2019. PMID: 31019051 Free PMC article.
-
Latent Epstein-Barr virus infection collaborates with Myc over-expression in normal human B cells to induce Burkitt-like Lymphomas in mice.PLoS Pathog. 2024 Apr 15;20(4):e1012132. doi: 10.1371/journal.ppat.1012132. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38620028 Free PMC article.
-
Epstein-Barr Virus B Cell Growth Transformation: The Nuclear Events.Viruses. 2023 Mar 24;15(4):832. doi: 10.3390/v15040832. Viruses. 2023. PMID: 37112815 Free PMC article. Review.
-
EBNA2 and Its Coactivator EBNA-LP.Curr Top Microbiol Immunol. 2015;391:35-59. doi: 10.1007/978-3-319-22834-1_2. Curr Top Microbiol Immunol. 2015. PMID: 26428371 Review.
Cited by
-
Continuous MYD88 Activation Is Associated With Expansion and Then Transformation of IgM Differentiating Plasma Cells.Front Immunol. 2021 May 4;12:641692. doi: 10.3389/fimmu.2021.641692. eCollection 2021. Front Immunol. 2021. PMID: 34017329 Free PMC article.
-
The viral and cellular microRNA targetome in lymphoblastoid cell lines.PLoS Pathog. 2012 Jan;8(1):e1002484. doi: 10.1371/journal.ppat.1002484. Epub 2012 Jan 26. PLoS Pathog. 2012. PMID: 22291592 Free PMC article.
-
Different patterns of Epstein-Barr virus latency in endemic Burkitt lymphoma (BL) lead to distinct variants within the BL-associated gene expression signature.J Virol. 2013 Mar;87(5):2882-94. doi: 10.1128/JVI.03003-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269792 Free PMC article.
-
Epstein-Barr virus oncoprotein super-enhancers control B cell growth.Cell Host Microbe. 2015 Feb 11;17(2):205-16. doi: 10.1016/j.chom.2014.12.013. Epub 2015 Jan 29. Cell Host Microbe. 2015. PMID: 25639793 Free PMC article.
-
Small molecule growth inhibitors of human oncogenic gammaherpesvirus infected B-cells.Mol Oncol. 2015 Feb;9(2):365-76. doi: 10.1016/j.molonc.2014.09.006. Epub 2014 Sep 26. Mol Oncol. 2015. PMID: 25306391 Free PMC article.
References
-
- Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, R. Levy, W. Wilson, M. R. Grever, J. C. Byrd, D. Botstein, P. O. Brown, and L. M. Staudt. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403503-511. - PubMed
-
- Baran-Marszak, F., J. Feuillard, I. Najjar, C. Le Clorennec, J. M. Bechet, I. Dusanter-Fourt, G. W. Bornkamm, M. Raphael, and R. Fagard. 2004. Differential roles of STAT1α and STAT1β in fludarabine-induced cell cycle arrest and apoptosis in human B cells. Blood 1042475-2483. - PubMed
-
- Bornkamm, G. W., C. Berens, C. Kuklik-Roos, J. M. Bechet, G. Laux, J. Bachl, M. Korndoerfer, M. Schlee, M. Holzel, A. Malamoussi, R. D. Chapman, F. Nimmerjahn, J. Mautner, W. Hillen, H. Bujard, and J. Feuillard. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33e137. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

