Engineering transcription factors with novel DNA-binding specificity using comparative genomics
- PMID: 19264798
- PMCID: PMC2677863
- DOI: 10.1093/nar/gkp079
Engineering transcription factors with novel DNA-binding specificity using comparative genomics
Abstract
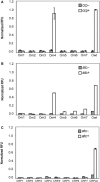
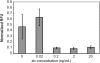
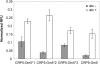
The transcriptional program for a gene consists of the promoter necessary for recruiting RNA polymerase along with neighboring operator sites that bind different activators and repressors. From a synthetic biology perspective, if the DNA-binding specificity of these proteins can be changed, then they can be used to reprogram gene expression in cells. While many experimental methods exist for generating such specificity-altering mutations, few computational approaches are available, particularly in the case of bacterial transcription factors. In a previously published computational study of nitrogen oxide metabolism in bacteria, a small number of amino-acid residues were found to determine the specificity within the CRP (cAMP receptor protein)/FNR (fumarate and nitrate reductase regulatory protein) family of transcription factors. By analyzing how these amino acids vary in different regulators, a simple relationship between the identity of these residues and their target DNA-binding sequence was constructed. In this article, we experimentally tested whether this relationship could be used to engineer novel DNA-protein interactions. Using Escherichia coli CRP as a template, we tested eight designs based on this relationship and found that four worked as predicted. Collectively, these results in this work demonstrate that comparative genomics can inform the design of bacterial transcription factors.
Figures



Similar articles
-
The Pseudomonas aeruginosa DNR transcription factor: light and shade of nitric oxide-sensing mechanisms.Biochem Soc Trans. 2011 Jan;39(1):294-8. doi: 10.1042/BST0390294. Biochem Soc Trans. 2011. PMID: 21265791
-
Transcriptional activation by FNR and CRP: reciprocity of binding-site recognition.Mol Microbiol. 1997 Feb;23(4):835-45. doi: 10.1046/j.1365-2958.1997.2811637.x. Mol Microbiol. 1997. PMID: 9157253
-
Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs.FEMS Microbiol Rev. 2003 Dec;27(5):559-92. doi: 10.1016/S0168-6445(03)00066-4. FEMS Microbiol Rev. 2003. PMID: 14638413 Review.
-
Transcription activation by FNR: evidence for a functional activating region 2.J Bacteriol. 2002 Nov;184(21):5855-61. doi: 10.1128/JB.184.21.5855-5861.2002. J Bacteriol. 2002. PMID: 12374818 Free PMC article.
-
FNR and its role in oxygen-regulated gene expression in Escherichia coli.FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review.
Cited by
-
Scaling up genetic circuit design for cellular computing: advances and prospects.Nat Comput. 2018;17(4):833-853. doi: 10.1007/s11047-018-9715-9. Epub 2018 Oct 5. Nat Comput. 2018. PMID: 30524216 Free PMC article.
-
Bayesian multiple-instance motif discovery with BAMBI: inference of recombinase and transcription factor binding sites.Nucleic Acids Res. 2011 Nov;39(21):e146. doi: 10.1093/nar/gkr745. Epub 2011 Sep 24. Nucleic Acids Res. 2011. PMID: 21948794 Free PMC article.
-
Modular control of multiple pathways using engineered orthogonal T7 polymerases.Nucleic Acids Res. 2012 Sep 1;40(17):8773-81. doi: 10.1093/nar/gks597. Epub 2012 Jun 28. Nucleic Acids Res. 2012. PMID: 22743271 Free PMC article.
-
Comparative Analysis of the IclR-Family of Bacterial Transcription Factors and Their DNA-Binding Motifs: Structure, Positioning, Co-Evolution, Regulon Content.Front Microbiol. 2021 Jun 10;12:675815. doi: 10.3389/fmicb.2021.675815. eCollection 2021. Front Microbiol. 2021. PMID: 34177859 Free PMC article.
-
Gly184 of the Escherichia coli cAMP receptor protein provides optimal context for both DNA binding and RNA polymerase interaction.J Microbiol. 2017 Oct;55(10):816-822. doi: 10.1007/s12275-017-7266-x. Epub 2017 Sep 28. J Microbiol. 2017. PMID: 28956357
References
-
- Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. - PubMed
-
- Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. - PubMed
-
- Rhodes D, Schwabe JW, Chapman L, Fairall L. Towards an understanding of protein-DNA recognition. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1996;351:501–509. - PubMed
-
- Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. - PubMed
-
- Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu. Rev. Biomed. Eng. 2003;5:179–206. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

