The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription
- PMID: 19264802
- PMCID: PMC2677877
- DOI: 10.1093/nar/gkp122
The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription
Abstract
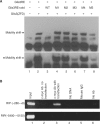
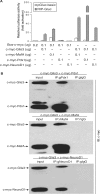
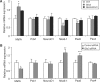
Glis3 is a member of the Krüppel-like family of transcription factors and is highly expressed in islet beta cells. Mutations in GLIS3 cause the syndrome of neonatal diabetes and congenital hypothyroidism (NDH). Our aim was to examine the role of Glis3 in beta cells, specifically with regard to regulation of insulin gene transcription. We demonstrate that insulin 2 (Ins2) mRNA expression in rat insulinoma 832/13 cells is markedly increased by wild-type Glis3 overexpression, but not by the NDH1 mutant. Furthermore, expression of both Ins1 and Ins2 mRNA is downregulated when Glis3 is knocked down by siRNA. Glis3 binds to the Ins2 promoter in the cell, detected by chromatin immunoprecipitation. Deletion analysis of Ins2 promoter identifies a sequence (5'-GTCCCCTGCTGTGAA-3') from -255 to -241 as the Glis3 response element and binding occur specifically via the Glis3 zinc finger region as revealed by mobility shift assays. Moreover, Glis3 physically and functionally interacts with Pdx1, MafA and NeuroD1 to modulate Ins2 promoter activity. Glis3 also may indirectly affect insulin promoter activity through upregulation of MafA and downregulation of Nkx6-1. This study uncovers a role of Glis3 for regulation of insulin gene expression and expands our understanding of its role in the beta cell.
Figures



References
-
- Kim YS, Lewandoski M, Perantoni AO, Kurebayashi S, Nakanishi G, Jetten AM. Identification of Glis1, a novel Gli-related, Kruppel-like zinc finger protein containing transactivation and repressor functions. J. Biol. Chem. 2002;277:30901–30913. - PubMed
-
- Zhang F, Nakanishi G, Kurebayashi S, Yoshino K, Perantoni A, Kim YS, Jetten AM. Characterization of Glis2, a novel gene encoding a Gli-related, Kruppel-like transcription factor with transactivation and repressor functions – roles in kidney development and neurogenesis. J. Biol. Chem. 2002;277:10139–10149. - PubMed
-
- Attanasio M, Uhlenhaut NH, Sousa VH, O’Toole JF, Otto E, Anlag K, Klugmann C, Treier AC, Helou J, Sayer JA, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat. Genet. 2007;39:1018–1024. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

