Metagenomic pyrosequencing and microbial identification
- PMID: 19264858
- PMCID: PMC2892905
- DOI: 10.1373/clinchem.2008.107565
Metagenomic pyrosequencing and microbial identification
Abstract
Background: The Human Microbiome Project has ushered in a new era for human metagenomics and high-throughput next-generation sequencing strategies.
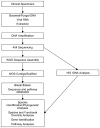
Content: This review describes evolving strategies in metagenomics, with a special emphasis on the core technology of DNA pyrosequencing. The challenges of microbial identification in the context of microbial populations are discussed. The development of next-generation pyrosequencing strategies and the technical hurdles confronting these methodologies are addressed. Bioinformatics-related topics include taxonomic systems, sequence databases, sequence-alignment tools, and classifiers. DNA sequencing based on 16S rRNA genes or entire genomes is summarized with respect to potential pyrosequencing applications.
Summary: Both the approach of 16S rDNA amplicon sequencing and the whole-genome sequencing approach may be useful for human metagenomics, and numerous bioinformatics tools are being deployed to tackle such vast amounts of microbiological sequence diversity. Metagenomics, or genetic studies of microbial communities, may ultimately contribute to a more comprehensive understanding of human health, disease susceptibilities, and the pathophysiology of infectious and immune-mediated diseases.
Figures


References
-
- Wilson M. Bacteriology of Humans: An Ecological Perspective. Malden, MA: Blackwell Publishing; 2008. pp. 266–326.
-
- Thies FL, Konig W, Konig B. Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J Med Microbiol. 2007;56:755–61. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

