Bif-1/endophilin B1: a candidate for crescent driving force in autophagy
- PMID: 19265852
- PMCID: PMC2697278
- DOI: 10.1038/cdd.2009.19
Bif-1/endophilin B1: a candidate for crescent driving force in autophagy
Abstract
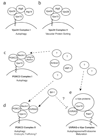
Autophagy is an intracellular bulk degradation system that plays a vital role in maintaining cellular homeostasis. This degradation process involves dynamic membrane rearrangements resulting in the formation of double-membraned autophagosomes. However, the driving force for generating curvature and deformation of isolation membranes remains a mystery. Bax-interacting factor 1 (Bif-1), also known as SH3GLB1 or Endophilin B1, was originally discovered as a Bax-binding protein. Bif-1 contains an amino-terminal N-BAR (Bin-Amphiphysin-Rvs) domain and a carboxy-terminal SH3 (Src-homology 3) domain and shows membrane binding and bending activities. It has been shown that Beclin1 is involved in the nucleation of autophagosomal membranes through an unknown mechanism. It is interesting that, Bif-1 forms a complex with Beclin1 through ultraviolet irradiation resistant-associated gene (UVRAG) and promotes the activation of the class III PI3 kinase, Vps34, in mammalian cells. In response to nutrient starvation, Bif-1 accumulates in punctate foci where it co-localizes with LC3, Atg5, and Atg9. Furthermore, Bif-1-positive, crescent-shaped small vesicles expand by recruiting and fusing with Atg9-positive small membranes to complete autophagosome formation. This review highlights the role of Bif-1 in the regulation of autophagy and discusses the potential involvement of Bif-1 in the biogenesis of membranes for the formation of autophagosomes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

