Rapid pathway evolution facilitated by horizontal gene transfers across prokaryotic lineages
- PMID: 19266023
- PMCID: PMC2644373
- DOI: 10.1371/journal.pgen.1000402
Rapid pathway evolution facilitated by horizontal gene transfers across prokaryotic lineages
Abstract
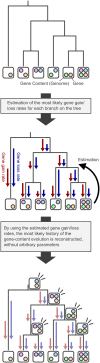
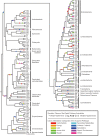
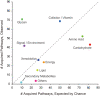
The evolutionary history of biological pathways is of general interest, especially in this post-genomic era, because it may provide clues for understanding how complex systems encoded on genomes have been organized. To explain how pathways can evolve de novo, some noteworthy models have been proposed. However, direct reconstruction of pathway evolutionary history both on a genomic scale and at the depth of the tree of life has suffered from artificial effects in estimating the gene content of ancestral species. Recently, we developed an algorithm that effectively reconstructs gene-content evolution without these artificial effects, and we applied it to this problem. The carefully reconstructed history, which was based on the metabolic pathways of 160 prokaryotic species, confirmed that pathways have grown beyond the random acquisition of individual genes. Pathway acquisition took place quickly, probably eliminating the difficulty in holding genes during the course of the pathway evolution. This rapid evolution was due to massive horizontal gene transfers as gene groups, some of which were possibly operon transfers, which would convey existing pathways but not be able to generate novel pathways. To this end, we analyzed how these pathways originally appeared and found that the original acquisition of pathways occurred more contemporaneously than expected across different phylogenetic clades. As a possible model to explain this observation, we propose that novel pathway evolution may be facilitated by bidirectional horizontal gene transfers in prokaryotic communities. Such a model would complement existing pathway evolution models.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Huynen MA, Gabaldon T, Snel B. Variation and evolution of biomolecular systems: searching for functional relevance. FEBS Lett. 2005;579:1839–1845. - PubMed
-
- Rison SC, Thornton JM. Pathway evolution, structurally speaking. Curr Opin Struct Biol. 2002;12:374–382. - PubMed
-
- Schmidt S, Sunyaev S, Bork P, Dandekar T. Metabolites: a helping hand for pathway evolution? Trends Biochem Sci. 2003;28:336–341. - PubMed
-
- Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

