The SET complex acts as a barrier to autointegration of HIV-1
- PMID: 19266025
- PMCID: PMC2644782
- DOI: 10.1371/journal.ppat.1000327
The SET complex acts as a barrier to autointegration of HIV-1
Abstract
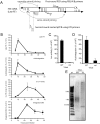
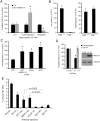
Retroviruses and retrotransposons are vulnerable to a suicidal pathway known as autointegration, which occurs when the 3'-ends of the reverse transcript are activated by integrase and then attack sites within the viral DNA. Retroelements have diverse strategies for suppressing autointegration, but how HIV-1 protects itself from autointegration is not well-understood. Here we show that knocking down any of the components of the SET complex, an endoplasmic reticulum-associated complex that contains 3 DNases (the base excision repair endonuclease APE1, 5'-3' exonuclease TREX1, and endonuclease NM23-H1), inhibits HIV-1 and HIV-2/SIV, but not MLV or ASV, infection. Inhibition occurs at a step in the viral life cycle after reverse transcription but before chromosomal integration. Antibodies to SET complex proteins capture HIV-1 DNA in the cytoplasm, suggesting a direct interaction between the SET complex and the HIV preintegration complex. Cloning of HIV integration sites in cells with knocked down SET complex components revealed an increase in autointegration, which was verified using a novel semi-quantitative nested PCR assay to detect autointegrants. When SET complex proteins are knocked down, autointegration increases 2-3-fold and chromosomal integration correspondingly decreases approximately 3-fold. Therefore, the SET complex facilitates HIV-1 infection by preventing suicidal autointegration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5:253–263. - PubMed
-
- Suzuki Y, Craigie R. The road to chromatin–nuclear entry of retroviruses. Nat Rev Microbiol. 2007;5:187–196. - PubMed
-
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. - PubMed
-
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

