Death and resurrection of the human IRGM gene
- PMID: 19266026
- PMCID: PMC2644816
- DOI: 10.1371/journal.pgen.1000403
Death and resurrection of the human IRGM gene
Abstract
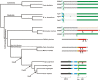
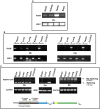
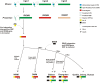
Immunity-related GTPases (IRG) play an important role in defense against intracellular pathogens. One member of this gene family in humans, IRGM, has been recently implicated as a risk factor for Crohn's disease. We analyzed the detailed structure of this gene family among primates and showed that most of the IRG gene cluster was deleted early in primate evolution, after the divergence of the anthropoids from prosimians ( about 50 million years ago). Comparative sequence analysis of New World and Old World monkey species shows that the single-copy IRGM gene became pseudogenized as a result of an Alu retrotransposition event in the anthropoid common ancestor that disrupted the open reading frame (ORF). We find that the ORF was reestablished as a part of a polymorphic stop codon in the common ancestor of humans and great apes. Expression analysis suggests that this change occurred in conjunction with the insertion of an endogenous retrovirus, which altered the transcription initiation, splicing, and expression profile of IRGM. These data argue that the gene became pseudogenized and was then resurrected through a series of complex structural events and suggest remarkable functional plasticity where alleles experience diverse evolutionary pressures over time. Such dynamism in structure and evolution may be critical for a gene family locked in an arms race with an ever-changing repertoire of intracellular parasites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, et al. Two families of GTPases dominate the complex cellular response to interferon-g. J Immunol. 1998;161:6715–6723. - PubMed
-
- Taylor GA. p47 GTPases: regulators of immunity to intracellular pathogens. Nature Reviews Immunology. 2004;4:100–109. - PubMed
-
- Howard J. The IRG proteins: A function in search of a mechanism. Immunobiology. 2008;213:367–375. - PubMed
-
- MacMicking J, Taylor GA, McKinney J. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

