Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment
- PMID: 19266028
- PMCID: PMC2644819
- DOI: 10.1371/journal.pgen.1000397
Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment
Abstract
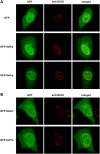
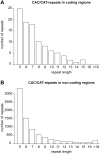
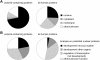
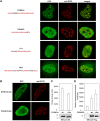
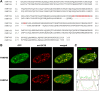
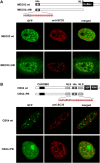
Single amino acid repeats are prevalent in eukaryote organisms, although the role of many such sequences is still poorly understood. We have performed a comprehensive analysis of the proteins containing homopolymeric histidine tracts in the human genome and identified 86 human proteins that contain stretches of five or more histidines. Most of them are endowed with DNA- and RNA-related functions, and, in addition, there is an overrepresentation of proteins expressed in the brain and/or nervous system development. An analysis of their subcellular localization shows that 15 of the 22 nuclear proteins identified accumulate in the nuclear subcompartment known as nuclear speckles. This localization is lost when the histidine repeat is deleted, and significantly, closely related paralogous proteins without histidine repeats also fail to localize to nuclear speckles. Hence, the histidine tract appears to be directly involved in targeting proteins to this compartment. The removal of DNA-binding domains or treatment with RNA polymerase II inhibitors induces the re-localization of several polyhistidine-containing proteins from the nucleoplasm to nuclear speckles. These findings highlight the dynamic relationship between sites of transcription and nuclear speckles. Therefore, we define the histidine repeats as a novel targeting signal for nuclear speckles, and we suggest that these repeats are a way of generating evolutionary diversification in gene duplicates. These data contribute to our better understanding of the physiological role of single amino acid repeats in proteins.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Huntley MA, Golding GB. Simple sequences are rare in the Protein Data Bank. Proteins. 2002;48:134–140. - PubMed
-
- Mar Alba M, Santibanez-Koref MF, Hancock JM. Amino acid reiterations in yeast are overrepresented in particular classes of proteins and show evidence of a slippage-like mutational process. J Mol Evol. 1999;49:789–797. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

