Cell-permeable beta-peptide inhibitors of p53/hDM2 complexation
- PMID: 19266537
- PMCID: PMC2995991
- DOI: 10.1002/cbic.200900049
Cell-permeable beta-peptide inhibitors of p53/hDM2 complexation
Abstract
Look at what the cat(ionic motif) dragged in! We report a general strategy to increase the cell permeability of beta(3)-peptides. Introduction of a minimal cationic motif within the folded structure of a high-affinity beta(3)-peptide ligand for hDM2 led to molecules with high 3(14)-helical structure, high hDM2 affinity and sufficient cell permeability to upregulate p53-dependent genes in live mammalian cells. Minimally cationic beta(3)-peptides represent the critical first step towards a class of protease-resistant peptidomimetics that might modulate intracellular biological pathways.
Figures

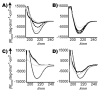
 (a) β53-12R2
(a) β53-12R2  , β53-12R3
, β53-12R3  , β53-8
, β53-8  ,β53-8R3
,β53-8R3  ; (b) β53-12SB2
; (b) β53-12SB2  , β53-12SB3
, β53-12SB3  , β53-3SB3
, β53-3SB3  ; (c) β53-12R6-1
; (c) β53-12R6-1  , β53-12R6-2
, β53-12R6-2  , βNEGR6
, βNEGR6  ; (d) β53-12R8
; (d) β53-12R8  , β53-3R8
, β53-3R8  . [θ]MRW = mean residue molar ellipticity.
. [θ]MRW = mean residue molar ellipticity.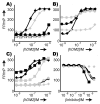
 (a) β53-12R2
(a) β53-12R2  , β53-12R3
, β53-12R3  , β53-8
, β53-8  ,β53-8R3
,β53-8R3  ; (b) β53-12SB2
; (b) β53-12SB2  , β53-12SB3
, β53-12SB3  , β53-3SB3
, β53-3SB3  ; (c) β53-12R6-1
; (c) β53-12R6-1  , β53-12R6-2
, β53-12R6-2  , βNEGR6
, βNEGR6  , β53-12R8
, β53-12R8  , β53-3R8
, β53-3R8  ; (d): Plot illustrating the observed polarization of the p53AD15-31flu complex with hDM21-188 as a function of the concentration of unlabeled peptide shown: β53-12, β53-12SB2, β53-12SB3, β53-12R8, and β53-3R8 (colors as above).
; (d): Plot illustrating the observed polarization of the p53AD15-31flu complex with hDM21-188 as a function of the concentration of unlabeled peptide shown: β53-12, β53-12SB2, β53-12SB3, β53-12R8, and β53-3R8 (colors as above).

References
-
- Seebach D, Overhand M, Kuhnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Helv Chim Acta. 1996;79:913–941.
-
- Cheng RP, Gellman SH, DeGrado WF. Chem Rev. 2001;101:3219–3232. - PubMed
-
- Gademann K, Kimmerlin T, Hoyer D, Seebach D. J Med Chem. 2001;44:2460–2468. - PubMed
-
- Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J Am Chem Soc. 2004;126:9468–9469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

