Retinal conformation and dynamics in activation of rhodopsin illuminated by solid-state H NMR spectroscopy
- PMID: 19267870
- PMCID: PMC2858981
- DOI: 10.1111/j.1751-1097.2008.00510.x
Retinal conformation and dynamics in activation of rhodopsin illuminated by solid-state H NMR spectroscopy
Abstract
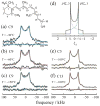
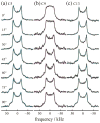
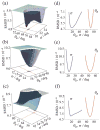
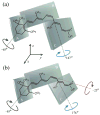
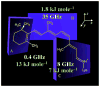
Solid-state NMR spectroscopy gives a powerful avenue for investigating G protein-coupled receptors and other integral membrane proteins in a native-like environment. This article reviews the use of solid-state (2)H NMR to study the retinal cofactor of rhodopsin in the dark state as well as the meta I and meta II photointermediates. Site-specific (2)H NMR labels have been introduced into three regions (methyl groups) of retinal that are crucially important for the photochemical function of rhodopsin. Despite its phenomenal stability (2)H NMR spectroscopy indicates retinal undergoes rapid fluctuations within the protein binding cavity. The spectral lineshapes reveal the methyl groups spin rapidly about their three-fold (C(3)) axes with an order parameter for the off-axial motion of SC(3) approximately 0.9. For the dark state, the (2)H NMR structure of 11-cis-retinal manifests torsional twisting of both the polyene chain and the beta-ionone ring due to steric interactions of the ligand and the protein. Retinal is accommodated within the rhodopsin binding pocket with a negative pretwist about the C11=C12 double bond. Conformational distortion explains its rapid photochemistry and reveals the trajectory of the 11-cis to trans isomerization. In addition, (2)H NMR has been applied to study the retinylidene dynamics in the dark and light-activated states. Upon isomerization there are drastic changes in the mobility of all three methyl groups. The relaxation data support an activation mechanism whereby the beta-ionone ring of retinal stays in nearly the same environment, without a large displacement of the ligand. Interactions of the beta-ionone ring and the retinylidene Schiff base with the protein transmit the force of the retinal isomerization. Solid-state (2)H NMR thus provides information about the flow of energy that triggers changes in hydrogen-bonding networks and helix movements in the activation mechanism of the photoreceptor.
Figures
References
-
- Pebay-Peyroula E, Rummel G, Rosenbusch JP, Landau EM. X-ray structure of bacteriorhodopsin at 2.5 Angstroms from microcrystals grown in lipid cubic phases. Science. 1997;277:1676–1681. - PubMed
-
- Chang G, Spencer RH, Lee AT, Barclay MT, Rees DC. Structure of the MscL homolog from Mycobacterium tuberculosis: A gated mechanosensitive ion channel. Science. 1998;282:2220–2226. - PubMed
-
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. - PubMed
-
- Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

