CNV-seq, a new method to detect copy number variation using high-throughput sequencing
- PMID: 19267900
- PMCID: PMC2667514
- DOI: 10.1186/1471-2105-10-80
CNV-seq, a new method to detect copy number variation using high-throughput sequencing
Abstract
Background: DNA copy number variation (CNV) has been recognized as an important source of genetic variation. Array comparative genomic hybridization (aCGH) is commonly used for CNV detection, but the microarray platform has a number of inherent limitations.
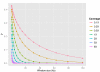
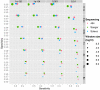
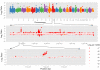
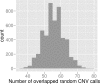
Results: Here, we describe a method to detect copy number variation using shotgun sequencing, CNV-seq. The method is based on a robust statistical model that describes the complete analysis procedure and allows the computation of essential confidence values for detection of CNV. Our results show that the number of reads, not the length of the reads is the key factor determining the resolution of detection. This favors the next-generation sequencing methods that rapidly produce large amount of short reads.
Conclusion: Simulation of various sequencing methods with coverage between 0.1x to 8x show overall specificity between 91.7 - 99.9%, and sensitivity between 72.2 - 96.5%. We also show the results for assessment of CNV between two individual human genomes.
Figures


References
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Månér S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. - DOI - PubMed
-
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK, Dallaire S, Freeman JL, González JR, Gratacòs M, Huang J, Kalaitzopoulos D, Komura D, MacDonald JR, Marshall CR, Mei R, Montgomery L, Nishimura K, Okamura K, Shen F, Somerville MJ, Tchinda J, Valsesia A, Woodwark C, Yang F, Zhang J, Zerjal T, Zhang J, Armengol L, Conrad DF, Estivill X, Tyler-Smith C, Carter NP, Aburatani H, Lee C, Jones KW, Scherer SW, Hurles ME. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. - DOI - PMC - PubMed
-
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Döhner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. doi: 10.1002/(SICI)1098-2264(199712)20:4<399::AID-GCC12>3.0.CO;2-I. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

