The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression
- PMID: 19267902
- PMCID: PMC2662859
- DOI: 10.1186/1471-2229-9-26
The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression
Abstract
Background: Lignin is a phenolic heteropolymer in secondary cell walls that plays a major role in the development of plants and their defense against pathogens. The biosynthesis of monolignols, which represent the main component of lignin involves many enzymes. The cinnamyl alcohol dehydrogenase (CAD) is a key enzyme in lignin biosynthesis as it catalyzes the final step in the synthesis of monolignols. The CAD gene family has been studied in Arabidopsis thaliana, Oryza sativa and partially in Populus. This is the first comprehensive study on the CAD gene family in woody plants including genome organization, gene structure, phylogeny across land plant lineages, and expression profiling in Populus.
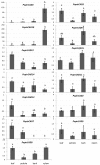
Results: The phylogenetic analyses showed that CAD genes fall into three main classes (clades), one of which is represented by CAD sequences from gymnosperms and angiosperms. The other two clades are represented by sequences only from angiosperms. All Populus CAD genes, except PoptrCAD 4 are distributed in Class II and Class III. CAD genes associated with xylem development (PoptrCAD 4 and PoptrCAD 10) belong to Class I and Class II. Most of the CAD genes are physically distributed on duplicated blocks and are still in conserved locations on the homeologous duplicated blocks. Promoter analysis of CAD genes revealed several motifs involved in gene expression modulation under various biological and physiological processes. The CAD genes showed different expression patterns in poplar with only two genes preferentially expressed in xylem tissues during lignin biosynthesis.
Conclusion: The phylogeny of CAD genes suggests that the radiation of this gene family may have occurred in the early ancestry of angiosperms. Gene distribution on the chromosomes of Populus showed that both large scale and tandem duplications contributed significantly to the CAD gene family expansion. The duplication of several CAD genes seems to be associated with a genome duplication event that happened in the ancestor of Salicaceae. Phylogenetic analyses associated with expression profiling and results from previous studies suggest that CAD genes involved in wood development belong to Class I and Class II. The other CAD genes from Class II and Class III may function in plant tissues under biotic stresses. The conservation of most duplicated CAD genes, the differential distribution of motifs in their promoter regions, and the divergence of their expression profiles in various tissues of Populus plants indicate that genes in the CAD family have evolved tissue-specialized expression profiles and may have divergent functions.
Figures



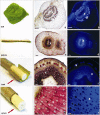
References
-
- Donaldson L, Hague J, Snell R. Lignin Distribution in Coppice Poplar, Linseed and Wheat Straw. Holzforschung. 2001;55:379–385. doi: 10.1515/HF.2001.063. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

