A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines
- PMID: 19267930
- PMCID: PMC2670824
- DOI: 10.1186/1471-2156-10-12
A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines
Abstract
Background: ENU-mutagenesis is a powerful technique to identify genes regulating mammalian development. To functionally annotate the distal region of mouse chromosome 4, we performed an ENU-mutagenesis screen using a balancer chromosome targeted to this region of the genome.
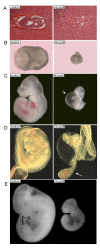
Results: We isolated 11 lethal lines that map to the region of chromosome 4 between D4Mit117 and D4Mit281. These lines form 10 complementation groups. The majority of lines die during embryonic development between E5.5 and E12.5 and display defects in gastrulation, cardiac development, and craniofacial development. One line displayed postnatal lethality and neurological defects, including ataxia and seizures.
Conclusion: These eleven mutants allow us to query gene function within the distal region of mouse chromosome 4 and demonstrate that new mouse models of mammalian developmental defects can easily and quickly be generated and mapped with the use of ENU-mutagenesis in combination with balancer chromosomes. The low number of mutations isolated in this screen compared with other balancer chromosome screens indicates that the functions of genes in different regions of the genome vary widely.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

