Oxidative stress alters base excision repair pathway and increases apoptotic response in apurinic/apyrimidinic endonuclease 1/redox factor-1 haploinsufficient mice
- PMID: 19268524
- PMCID: PMC2677124
- DOI: 10.1016/j.freeradbiomed.2009.02.021
Oxidative stress alters base excision repair pathway and increases apoptotic response in apurinic/apyrimidinic endonuclease 1/redox factor-1 haploinsufficient mice
Abstract
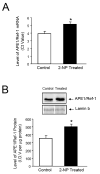
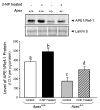
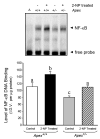
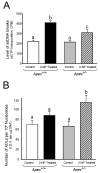
Apurinic/apyrimidinic endonuclease 1/redox factor-1 (APE1/Ref-1) is the redox regulator of multiple stress-inducible transcription factors, such as NF-kappaB, and the major 5'-endonuclease in base excision repair (BER). We utilized mice containing a heterozygous gene-targeted deletion of APE1/Ref-1 (Apex(+/-)) to determine the impact of APE1/Ref-1 haploinsufficiency on the processing of oxidative DNA damage induced by 2-nitropropane (2-NP) in the liver tissue of mice. APE1/Ref-1 haploinsufficiency results in a significant decline in NF-kappaB DNA-binding activity in response to oxidative stress in liver. In addition, loss of APE1/Ref-1 increases the apoptotic response to oxidative stress, in which significant increases in GADD45g expression, p53 protein stability, and caspase activity are observed. Oxidative stress displays a differential impact on monofunctional (UNG) and bifunctional (OGG1) DNA glycosylase-initiated BER in the liver of Apex(+/-) mice. APE1/Ref-1 haploinsufficiency results in a significant decline in the repair of oxidized bases (e.g., 8-OHdG), whereas removal of uracil is increased in liver nuclear extracts of mice using an in vitro BER assay. Apex(+/-) mice exposed to 2-NP displayed a significant decline in 3'-OH-containing single-strand breaks and an increase in aldehydic lesions in their liver DNA, suggesting an accumulation of repair intermediates of failed bifunctional DNA glycosylase-initiated BER.
Figures



Similar articles
-
The role of the N-terminal domain of human apurinic/apyrimidinic endonuclease 1, APE1, in DNA glycosylase stimulation.DNA Repair (Amst). 2018 Apr;64:10-25. doi: 10.1016/j.dnarep.2018.02.001. Epub 2018 Feb 11. DNA Repair (Amst). 2018. PMID: 29475157
-
Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress.Neuroscience. 2007 Apr 14;145(4):1201-12. doi: 10.1016/j.neuroscience.2006.10.010. Epub 2006 Nov 13. Neuroscience. 2007. PMID: 17101234
-
Apurinic/apyrimidinic endonuclease (APE/REF-1) haploinsufficient mice display tissue-specific differences in DNA polymerase beta-dependent base excision repair.J Biol Chem. 2004 Apr 30;279(18):18425-33. doi: 10.1074/jbc.M313983200. Epub 2004 Feb 18. J Biol Chem. 2004. PMID: 14973123
-
The DNA base excision repair protein Ape1/Ref-1 as a therapeutic and chemopreventive target.Mol Aspects Med. 2007 Jun-Aug;28(3-4):375-95. doi: 10.1016/j.mam.2007.04.005. Epub 2007 May 3. Mol Aspects Med. 2007. PMID: 17560642 Review.
-
Inhibitors of nuclease and redox activity of apurinic/apyrimidinic endonuclease 1/redox effector factor 1 (APE1/Ref-1).Bioorg Med Chem. 2017 May 1;25(9):2531-2544. doi: 10.1016/j.bmc.2017.01.028. Epub 2017 Jan 21. Bioorg Med Chem. 2017. PMID: 28161249 Review.
Cited by
-
Knockout and Inhibition of Ape1: Roles of Ape1 in Base Excision DNA Repair and Modulation of Gene Expression.Antioxidants (Basel). 2022 Sep 15;11(9):1817. doi: 10.3390/antiox11091817. Antioxidants (Basel). 2022. PMID: 36139891 Free PMC article.
-
Excessive Folic Acid Mimics Folate Deficiency in Human Lymphocytes.Curr Issues Mol Biol. 2022 Mar 23;44(4):1452-1462. doi: 10.3390/cimb44040097. Curr Issues Mol Biol. 2022. PMID: 35723355 Free PMC article.
-
A Novel Hypomorphic Apex1 Mouse Model Implicates Apurinic/Apyrimidinic Endonuclease 1 in Oxidative DNA Damage Repair in Gastric Epithelial Cells.Antioxid Redox Signal. 2023 Jan;38(1-3):183-197. doi: 10.1089/ars.2021.0119. Epub 2022 Aug 29. Antioxid Redox Signal. 2023. PMID: 35754343 Free PMC article.
-
Ubiquitin ligase UBR3 regulates cellular levels of the essential DNA repair protein APE1 and is required for genome stability.Nucleic Acids Res. 2012 Jan;40(2):701-11. doi: 10.1093/nar/gkr744. Epub 2011 Sep 20. Nucleic Acids Res. 2012. PMID: 21933813 Free PMC article.
-
Role of primary aging hallmarks in Alzheimer´s disease.Theranostics. 2023 Jan 1;13(1):197-230. doi: 10.7150/thno.79535. eCollection 2023. Theranostics. 2023. PMID: 36593969 Free PMC article. Review.
References
-
- Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. - PubMed
-
- Mullaart E, Lohman PHM, Berends F, Vijg J. DNA damage metabolism and aging. Mutat Res. 1990;237:189–210. - PubMed
-
- Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. - PubMed
-
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. - PubMed
-
- Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat Res. 2003;531:231–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

