Detection of gene loops by 3C in yeast
- PMID: 19269325
- PMCID: PMC2761752
- DOI: 10.1016/j.ymeth.2009.02.018
Detection of gene loops by 3C in yeast
Abstract
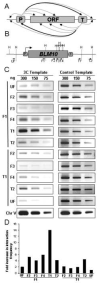
"Chromosome conformation capture" (3C) is a powerful method to detect physical interaction between any two genomic loci. 3C involves formaldehyde crosslinking to stabilize transient interactions, followed by restriction digestion, ligation and locus-specific PCR. Accordingly, 3C reveals complex three-dimensional interactions between distal genetic elements within intact cells at high resolution. Here, we describe a modified 3C protocol designed for detection of transient chromatin interactions in the yeast Saccharomyces cerevisiae. Using this protocol, we are able to detect juxtaposition of promoter and terminator regions of genes with ORFs as short as 1kb in length. We anticipate that this method will be generally applicable to detect dynamic, short-range chromatin interactions and will facilitate the characterization of gene loops and their functional consequences.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

