3D imaging of diatoms with ion-abrasion scanning electron microscopy
- PMID: 19269330
- PMCID: PMC2743382
- DOI: 10.1016/j.jsb.2009.02.014
3D imaging of diatoms with ion-abrasion scanning electron microscopy
Abstract
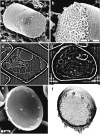
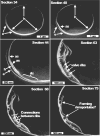
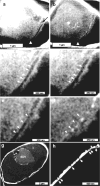
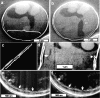
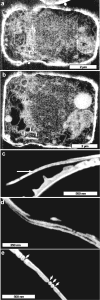
Ion-abrasion scanning electron microscopy (IASEM) takes advantage of focused ion beams to abrade thin sections from the surface of bulk specimens, coupled with SEM to image the surface of each section, enabling 3D reconstructions of subcellular architecture at approximately 30nm resolution. Here, we report the first application of IASEM for imaging a biomineralizing organism, the marine diatom Thalassiosira pseudonana. Diatoms have highly patterned silica-based cell wall structures that are unique models for the study and application of directed nanomaterials synthesis by biological systems. Our study provides new insights into the architecture and assembly principles of both the "hard" (siliceous) and "soft" (organic) components of the cell. From 3D reconstructions of developmentally synchronized diatoms captured at different stages, we show that both micro- and nanoscale siliceous structures can be visualized at specific stages in their formation. We show that not only are structures visualized in a whole-cell context, but demonstrate that fragile, early-stage structures are visible, and that this can be combined with elemental mapping in the exposed slice. We demonstrate that the 3D architectures of silica structures, and the cellular components that mediate their creation and positioning can be visualized simultaneously, providing new opportunities to study and manipulate mineral nanostructures in a genetically tractable system.
Figures





References
-
- Armbrust E, Berges J, Bowler C, Green B, Martinez D, Putnam N, Zhou S, Allen A, Apt K, Bechner M, Brzezinski M, Chaal B, Chiovitti A, Davis A, Demarest M, Detter J, Glavina T, Goodstein D, Hadi M, Hellsten U, Hildebrand M, Jenkins B, Jurka J, Kapitonov V, Kröger N, Lau W, Lane T, Larimer F, Lippmeier J, Lucas S, Medina M, Montsant A, Obornik M, Parker M, Palenik B, Pazour G, Richardson P, Rynearson T, Saito M, Schwartz D, Thamatrakoln K, Valentin K, Vardi A, Wilkerson F, Rokhsar D. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. - PubMed
-
- Badour SS. Experimental separation of cell division and silica shell formation in Cyclotella cryptica. Archiv fur Mikrobiol. 1968;62:17–33.
-
- Bao ZH, Weatherspoon MR, Shian S, Cai Y, Graham PD, Allan SM, Ahmad G, Dickerson MB, Church BC, Kang ZT, Abernathy HW, Summers CJ, Liu ML, Sandhage KH. Chemical reduction of three-dimensional silica micro-assemblies into microporous silicon replicas. Nature. 2007;446:172–175. - PubMed
-
- Bright D. Lispix: An image processing and analysis tool for the PC. 2007. http://www.nist.gov/lispix/doc/contents.htm.
-
- Darley WM, Volcani BE. Role of silicon in diatom metabolism: A silicon requirement for deoxyribonucleic acid synthesis in the diatom Cylindrotheca fusiformis Reimann and Lewin. Exp Cell Res. 1969;58:334–342. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

