The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner
- PMID: 19269976
- PMCID: PMC2673300
- DOI: 10.1074/jbc.M808850200
The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner
Abstract
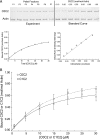
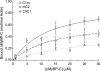
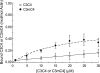
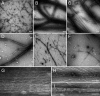
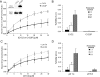
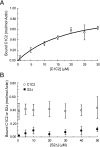
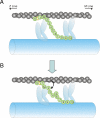
Cardiac myosin-binding protein C (cMyBP-C) is a regulatory protein expressed in cardiac sarcomeres that is known to interact with myosin, titin, and actin. cMyBP-C modulates actomyosin interactions in a phosphorylation-dependent way, but it is unclear whether interactions with myosin, titin, or actin are required for these effects. Here we show using cosedimentation binding assays, that the 4 N-terminal domains of murine cMyBP-C (i.e. C0-C1-m-C2) bind to F-actin with a dissociation constant (K(d)) of approximately 10 microm and a molar binding ratio (B(max)) near 1.0, indicating 1:1 (mol/mol) binding to actin. Electron microscopy and light scattering analyses show that these domains cross-link F-actin filaments, implying multiple sites of interaction with actin. Phosphorylation of the MyBP-C regulatory motif, or m-domain, reduced binding to actin (reduced B(max)) and eliminated actin cross-linking. These results suggest that the N terminus of cMyBP-C interacts with F-actin through multiple distinct binding sites and that binding at one or more sites is reduced by phosphorylation. Reversible interactions with actin could contribute to effects of cMyBP-C to increase cross-bridge cycling.
Figures
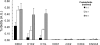

References
-
- Richard, P., Charron, P., Carrier, L., Ledeuil, C., Cheav, T., Pichereau, C., Benaiche, A., Isnard, R., Dubourg, O., Burban, M., Gueffet, J. P., Millaire, A., Desnos, M., Schwartz, K., Hainque, B., and Komajda, M. (2003) Circulation 107 2227-2232 - PubMed
-
- Dhandapany, P. S., Sadayappan, S., Xue, Y., Powell, G. T., Rani, D. S., Nallari, P., Rai, T. S., Khullar, M., Soares, P., Bahl, A., Tharkan, J. M., Vaideeswar, P., Rathinavel, A., Narasimhan, C., Ayapati, D. R., Ayub, Q., Mehdi, S. Q., Oppenheimer, S., Richards, M. B., Price, A. L., Patterson, N., Reich, D., Singh, L., Tyler-Smith, C., and Thangaraj, K. (2009) Nat. Genet. 41 187-191 - PMC - PubMed
-
- Harris, S. P., Bartley, C. R., Hacker, T. A., McDonald, K. S., Douglas, P. S., Greaser, M. L., Powers, P. A., and Moss, R. L. (2002) Circ. Res. 90 594-601 - PubMed
-
- Korte, F. S., McDonald, K. S., Harris, S. P., and Moss, R. L. (2003) Circ. Res. 93 752-758 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

