A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells
- PMID: 19270079
- PMCID: PMC2664065
- DOI: 10.1073/pnas.0900691106
A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells
Abstract
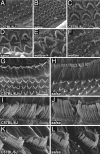
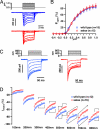
Deafness is the most common form of sensory impairment in humans and is frequently caused by single gene mutations. Interestingly, different mutations in a gene can cause syndromic and nonsyndromic forms of deafness, as well as progressive and age-related hearing loss. We provide here an explanation for the phenotypic variability associated with mutations in the cadherin 23 gene (CDH23). CDH23 null alleles cause deaf-blindness (Usher syndrome type 1D; USH1D), whereas missense mutations cause nonsyndromic deafness (DFNB12). In a forward genetic screen, we have identified salsa mice, which suffer from hearing loss due to a Cdh23 missense mutation modeling DFNB12. In contrast to waltzer mice, which carry a CDH23 null allele mimicking USH1D, hair cell development is unaffected in salsa mice. Instead, tip links, which are thought to gate mechanotransduction channels in hair cells, are progressively lost. Our findings suggest that DFNB12 belongs to a new class of disorder that is caused by defects in tip links. We propose that mutations in other genes that cause USH1 and nonsyndromic deafness may also have distinct effects on hair cell development and function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Comment in
-
Unraveling cadherin 23's role in development and mechanotransduction.Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):4959-60. doi: 10.1073/pnas.0902008106. Epub 2009 Mar 25. Proc Natl Acad Sci U S A. 2009. PMID: 19321743 Free PMC article. Review. No abstract available.
References
-
- Baux D, et al. UMD-USHbases: A comprehensive set of databases to record and analyse pathogenic mutations and unclassified variants in seven Usher syndrome causing genes. Hum Mutat. 2008;29:E76–E87. - PubMed
-
- Becirovic E, et al. Usher syndrome type 1 due to missense mutations on both CDH23 alleles: Investigation of mRNA splicing. Hum Mutat. 2008;29:452. - PubMed
-
- Bolz H, et al. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

